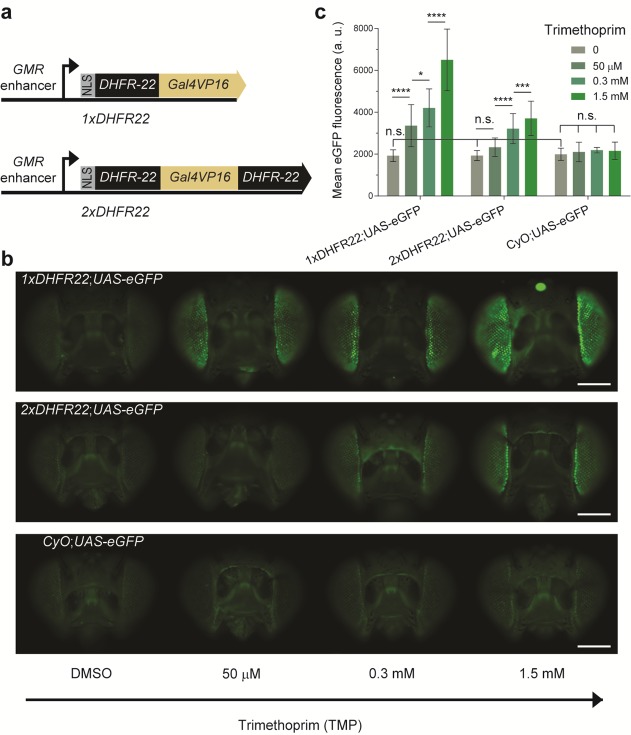
Figure 4.
The drug-stabilizable Gal4 variants 1xDHFR22 and 2xDHFR22 function in vivo in the format of the widely used Gal4-UAS bipartite expression system for Drosophila. (a) Schematic representation of the constructs used to create drug-inducible Gal4 driver lines. 1xDHFR22 encodes the single DHFR-DD architecture with the DHFR variant 22,28 as a fusion to the Gal4VP16 transcription factor. A nuclear localization signal (NLS) is added N-terminally and expression is driven by the eye-specific enhancer, glass multiple reporter (GMR). Similarly, 2xDHFR22 encodes the double architecture of DHFR22-DD. (b) A population of F1 progenies from 1xDHFR22 and 2xDHFR22 genetic crosses with UAS-eGFP reporter line was allowed to feed on standard fly food supplemented with DMSO (mock-treatment) or various concentrations of TMP for 5-days. A negative control population was derived from the Curly wings phenotype resulting from a dominant CyO marker from the heterozygote 1xDHFR or 2xDHFR driver line. Samples of the population were imaged by fluorescence microscopy. Representative images of adult fly eyes display an increase in eGFP fluorescence intensity as a function of the inducer TMP. The upper, middle and lower panels display, respectively, F1 progenies with genotypes 1xDHFR22;UAS-eGFP, 2xDHFR22;UAS-eGFP and CyO;UAS-eGFP. Scale bar: 1 mm. (c) Quantification of eGFP fluorescence intensity in the Drosophila adult eyes either mock-treated with DMSO or with various concentrations of TMP. Data are presented as the mean fluorescence detected per eye. The statistical significance resulting from a one-way ANOVA and Tukey’s post hoc test is summarized with asterisk marks representing the level of significance (n.s.= P-value > 0.05, * = P-value ≤ 0.05, *** = P-value ≤ 0. 001, and **** = P-value ≤ 0.0001) on the indicated data set. The error bars represent the standard deviation over the mean across the biological replicates (n = 8–76 individual eyes per dose).