Abstract
This article reports the first absolute quantitative analysis of expression patterns of murine transcripts (Gsta1/2, Gsta3, Gsta4, Gstm1, Gstm2, Gstm3, Gsto1, Gstp1/2, Gstt1, Gstt2) coding for most glutathione S-transferases (GSTs) of alpha, mu, omega, pi, and theta classes. We examine how the steady-state numbers of transcripts are modulated in association with: three animal organs (liver, kidney, and lung) where extensive detoxification occurs; two species (Mus musculus and Mus spretus) representing common laboratory and aboriginal mice; and two genetic and animal living conditions (wild-derived inbred animals and free-living mice). Moreover, quantitations performed examine how the pulmonary steady-state Gst mRNA amounts are affected in M. musculus by paraquat (a superoxide generator), and in M. spretus by dwelling at a polluted area. The results point to complex tissue-, species-, and life condition-dependent expression of the investigated transcripts. Among others, they show: i) the ubiquity of most transcripts, except Gstm3 mRNA that was virtually absent or at very low amounts (≤0.001 molecules/pg) in kidney and lung of M. spretus; ii) unique expression profiles for each transcript and mouse organ examined; iii) outstanding species-specific differences in basal amounts of most Gst mRNAs, this effect being most apparent in the case of Gsta1/2, Gsta3, Gstm2, Gsto1, Gstt1, and Gstt2; iv) paraquat-induced upregulation of most Gst mRNAs, with the notable exception of those coding for theta class GSTs; v) a tendency for mice dwelling at a wildlife reserve of having lower and more homogeneous Gsta3 mRNA levels than those collected in an anthropogenic environment.
Key words: Mus spretus, Gst transcript copy numbers, Biomarkers, Oxidative stress, Detoxification, Quantitative RT-PCR
GLUTATHIONE S-transferases (GSTs) are one of the most important multigene families of enzymes for the detoxification of xenobiotic and endogenous electrophilic compounds. They catalyze the conjugation of relatively hydrophobic electrophilic molecules with reduced glutathione (GSH), thereby enhancing their hydrophilicity and excretion. Key substrates of these phase II enzymes include epoxides derived from polycyclic aromatic hydrocarbons through the catalytic actions of phase I cytochrome P-450s, as well as numerous byproducts of oxidative stress (12). In addition to their traditional function in the conjugative detoxification pathway, members of this family are being implicated in a growing list of other pivotal cellular functions, for which GSTs are now being considered as multifunctional enzymes devoted to various aspects of cell defense (12,15).
There are two GST superfamilies, one soluble and mainly cytosolic and the other membrane bound. The soluble GSTs exist either as homo- or heterodimers, and have two domains per monomer, one for GSH and other for the electrophilic substrate (22). The larger superfamily of soluble GSTs is grouped into at least eight distinct classes (alpha, kappa, mu, omega, pi, sigma, theta, and zeta), on the basis of their immunological properties, substrate specificity, and amino acid sequences (12,15).
The mouse is the best known vertebrate model organism. Mus musculus is the source of most classical inbred laboratory mouse strains. The laboratory mouse is a good tool for understanding genetic influences of differences in phenotypes. However, genetic and phenotypic variations of the laboratory mouse are small compared with those of the wild mice. Mus spretus is the best characterized of the aboriginal species, which separated from the M. musculus group of house mouse subspecies more than 1 million years ago. M. spretus is known as a powerful genetic research tool and as an alternative to M. musculus for a wide variety of studies [e.g., see (1,6,13,23)]. Recently, M. spretus is being used as an indicator species in environmental pollution studies, through the use of different cytogenetic and biochemical biomarkers including total cytosolic and microsomal GST activities (4,21,25).
Quantitation of expression profiles of key genes in mammals is fundamental in toxicology, epidemiology, environmental health, and clinical medicine. For instance, profiling metabolic enzymes within tissues permits better prediction of sites of toxicity, metabolic intermediates, and responses to exposure to specific environmental pollutants. This investigation sought to establish the expression patterns of most (12/16) murine genes coding for GST enzymes of classes alpha, mu, omega, pi, and theta. Given that control of mRNA levels is crucial in regulating protein production in mammals and that modulation of transcription can only be fully understood in absolute quantitative terms, herein we provide for the first time valuable new information based on actual numbers of Gst mRNA molecules. To this end, we used a quantitatively rigorous approach based on reverse transcription followed by a combination of multiplex and realtime PCR amplification (14). We wanted to know how the organ context, the mouse species, and the environment modulate the absolute expression patterns of the investigated transcripts. Aside from the inherent value of the absolute expression patterns given here, data reported provide a novel picture of how a coordinated fine-tune of mRNA copy numbers might be important for the organism to respond effectively to diverse stimuli, therefore contributing to what remains to be done in the course of the pursuit of the medical benefits that should be derived from the study of the factors modulating the GST expression levels.
MATERIALS AND METHODS
Animals
M. musculus (inbred BALB/c strain; BALB/cByJ substrain) mice were from Charles River Laboratory (Spain). Wild-derived inbred M. spretus (SPRET/EiJ strain) mice were from Jackson Laboratory (USA). Wild M. spretus mice were captured with live Sherman traps as described previously (21). Animals were individually killed by cervical dislocation. The liver, kidney, and lung were removed, in that order, and immediately frozen in liquid nitrogen. Care was taken to ensure that a maximum of 5 min elapsed between time of death and harvest of the last organ. Wild mice were taken alive to the nearest laboratory at the Doñana Biological Reserve, where their sex and weight were determined. Immediately after, the organs were removed and taken frozen in liquid nitrogen to Córdoba University. Mice were handled according to the norms stipulated by the European Community. The investigation was performed after approval by the Ethical Committee of the University of Córdoba (Spain).
Treatments
To quantitate the abundance of Gst transcripts under an oxidizing condition, the series of lungs that was generated in a previous study in M. musculus (19) was examined. This series comprises organs from male BALB/cByJ of 7 weeks of age challenged with paraquat (PQ) at the moderate dose of 30 mg kg−1 body weight [approx. the LD10 dose (2)] for 30, 60, 120, and 240 min. This time course experiment was completed with a dose–response study in which male mice (BALB/cByJ, 7 weeks old) were stressed with 60 and 120 mg kg−1 body weight of PQ during 240 min.
RNA Preparations and Reverse Transcription
RNA extraction and synthesis of standard RNA and of cDNA were as detailed previously (19). RNA sample quality was checked electrophoretically, and quantification was done spectrophotometrically. Absence of genomic DNA contamination was checked by PCR amplification of RNA samples without prior cDNA synthesis.
Primers
Sequences of genes for primer design were obtained from GenBank™. Primers were made with the Oligo 6.1.1/98 (Molecular Biology Insights, Plymouth, MN, USA) program, and were located in the protein-coding region of the respective mRNAs. To obtain the highest specificity and performance, primers were required to have high T m (≥80°C) and optimal 3′ ΔG (≥−6.3 kcal/mol) values. Because of the restrictive parameters used in primer selection (20) and the extensive nucleotide sequence identity shared by the 12 Gst mRNAs investigated herein, the designed primers were distributed into two sets for multiplexed PCR (MPCR). Set A included primer pairs for amplification of Gstal/2, Gsta3, Gstm2, Gstpl/2, and Gsttl, whereas set B included primer pairs for amplification of Gsta4, Gstml, Gstm3, Gstol, and Gsttl. Two pairs of primers in set A do not discriminate between mRNAs coding for Gsta1 versus Gsta2 and Gstp1 versus Gstp2 isoforms, respectively. All designed primer pairs generate specific PCR products of the expected length and nucleotide sequence. Forward primers were labeled with 4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein for MPCR. Primer sequences are available from the authors on request.
M. spretus Gst Sequences
Most primers designed (based on M. musculus sequences) amplified in M. spretus single products exhibiting 100% nucleotide sequence identity with M. musculus. Exceptions were Gstml (A260 to G), Gstml (A438 to T, and T471 to C), Gstm3 (G 131 to A), Gstp1/2 (C501 to T, and C507 to A), and Gstt2 (G492 to A). Of these nucleotide differences, only two lead to changes in the deduced amino acid sequence: Asp87 to Gly (Gstm1) and Ser44 to Asn (Gstm3). The possible ultimate effects of such amino acid substitutions on the resulting GST protein have not been investigated so far. M. spretus nucleotide sequences have been submitted to GenBank™ under the following accession numbers: AY355150 (Gstal), AY355151 (Gstal), AY355152 (Gsta3), AY355153 (Gsta4), AY355154 (Gstml), AY355155 (Gstm3), AY355156 (Gstol), AY355157 (Gstp1/2), AY355158 (Gsttl), and AY355159 (Gsttl).
Real-Time and Multiplex PCR
Real-time PCRs were performed in quadruplicate as detailed previously (19). No primer dimers were detected, and the investigated transcripts showed optimal PCR efficiencies (from 0.98 to 1.00) in both mouse species. An absolute calibration curve was constructed with an external standard in the range of 102 to 109 RNA molecules. The number of mRNA molecules was calculated from the linear regression of the standard curve, as described previously (19). MPCR conditions (primer concentrations and PCR cycle number) were optimized for each primer set, mouse organ, and species as detailed previously (20), to ensure that all amplifications were in the exponential phase and the efficiencies remained constant in the course of the PCR. Components of the reaction mixture were as described previously (20). Relative quantitation of the MPCR products was as detailed previously (20). MPCR reactions were carried out in duplicate. Absolute measurements (mRNA molecules) were inferred from the relative expression ratios given by MPCR, as we have described recently (14). Thorough experimental conditions are available from the authors on request.
Statistical Analysis
Basal amounts of Gst transcripts were quantitated in the liver, kidney, and lung of each animal (n ≥ 5 mice). The Student’s t-test and the nonparametric Mann-Whitney test were used to detect differences in basal amounts of Gst transcripts. The amounts of Gst transcripts following PQ exposure were quantitated in the lung of each treated animal (n = 3 mice). Statistical significance was evaluated by using ANOVA followed by the Student-Newman-Keuls method. A Gst gene was considered responsive to PQ if the copy number of mRNA was statistically different from the corresponding control value (saline-injected animals) on at least two consecutive PQ doses and/or exposure times. Significant differences were determined at p < 0.05.
RESULTS
Absolute Quantitation of Basal Gst mRNA Levels
Basal expression profiles of Gst genes were determined in the liver, kidney, and lung of male mice (Table 1). BALB/c (M. musculus) is popular as a general purpose inbred strain for laboratory studies in many different research disciplines. Adult mice of the BALB/cByJ substrain were analyzed in this study. The Algerian mouse (M. spretus) is a nonprotected species that has been used as sentinel organism in several environmental studies (4,21). Here, both inbred and free-living M. spretus mice were analyzed. SPRET/EiJ is a wild-derived (geographic origin: Puerto Real, Cádiz, SW Spain) strain of M. spretus that has been maintained by sibling mating for 65 generations at the Jackson Laboratory (USA). Hence, these mice are as genetically alike as possible, being homozygous at virtually all of their loci. Wild M. spretus were captured at the Santa Olalla Lagoon (SOL). This sampling area, located at the core of the Doñana Biological Reserve (Huelva, SW Spain), is considered as nonpolluted according to several biochemical biomarkers (4,21). The Santa Olalla Lagoon is about 50 km NW Puerto Real.
TABLE 1.
MALE MICE USED FOR ABSOLUTE QUANTITATION OF STEADY-STATE Gst mRNA LEVELS
| Mouse | Age (Weeks) | Weight (g) |
|---|---|---|
| M. musculus (BALB/cByJ) | ||
| MM09 | 7 | 22.9 |
| MM10 | 7 | 26.8 |
| MM11 | 7 | 23.0 |
| MM12 | 7 | 25.0 |
| MM13 | 7 | 25.5 |
| Mean (±0.75 SEM) | 24.6 | |
| M. spretus (SPRET/EiJ) | ||
| MS01 | 8 | 11.0 |
| MS02 | 8 | 12.5 |
| MS03 | 8 | 12.4 |
| MS04 | 11 | 12.7 |
| MS05 | 11 | 13.1 |
| MS06 | 8 | 11.7 |
| Mean (±0.31 SEM) | 12.2 | |
| M. spretus (SOL) | ||
| SOL05 | unknown | 12.0 |
| SOL08 | unknown | 12.0 |
| SOL22 | unknown | 12.2 |
| SOL32 | unknown | 12.3 |
| SOL39 | unknown | 12.9 |
| Mean (±0.16 SEM) | 12.3 | |
The refined reverse transcription-PCR analysis conducted in this work allowed us to quantitate accurately the steady-state copy numbers of all examined Gst mRNAs. Both low (0.001 mRNA molecules/pg of total RNA) and high (more than 700 molecules/pg) basal expression levels were determined in a highly quantitative manner. All transcripts were ubiquitously expressed, except Gstm3 mRNA, which was virtually absent (<0.001 molecules/pg) in kidney from M. spretus. Data reported show that basal expression profiles differed substantially depending on the organ (Tables 2–4) and mouse species (Figs. 1–3).
TABLE 2.
STEADY-STATE LEVELS OF Gst mRNAs IN M. MUSCULUS (BALB/cByJ)
| Mouse | Gsta1/2 | Gsta3 | Gsta4 | Gstm1 | Gstm2 | Gstm3 | Gsto1 | Gstp1/2 | Gstt1 | Gstt2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Liver | 6.2 ± 1.1a | 120 ±9.9 | 13 ± 0.63b | 322 ± 28 | 45 ± 4.6c | 4.2 ± 1.3a,d | 12 ± 0.36b | 94 ± 4.4 | 48 ± 4.5c | 2.9 ± 0.41d |
| Kidney | 4.0 ± 0.83a | 10 ± 1.1e | 3.8 ± 0.33a | 159 ± 15 | 42 ± 2.7c | 0.005 ± 0.0005 | 7.0 ± 0.61 | 35 ± 2.1c,f | 29 ± 2.5f | 12 ± 1.6e |
| Lung | 0.60 ± 0.12g | 20 ± 2.7h,i | 12 ± 1.0b | 100 ± 8.5 | 68 ± 4.0 | 0.04 ± 0.004 | 12 ± 0.81b | 27 ± 2.2h | 19 ± 1.8i | 0.85 ± 0.06g |
Data are the means ± SEM of mRNA molecules per pg of total RNA. Basal amounts of different Gst mRNAs from the same organ as well as basal amounts of each Gst mRNA in the three examined mouse organs were compared. Average numbers with the same superscript are not statistically different (p > 0.05).
TABLE 4.
STEADY-STATE LEVELS OF Gst mRNAs IN WILD M. SPRETUS FROM SOL SAMPLING AREA
| Mouse | Gsta1/2 | Gsta3 | Gsta4 | Gstm1 | Gstm2 | Gstm3 | Gsto1 | Gstp1/2 | Gstt1 | Gstt2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Liver | 92 ± 16a | 0.11 ± 0.02 | 6.1 ± 1.3b | 276 ± 72c,d | 150 ± 26a,d | 0.07 ± 0.04 | 65 ± 1.3e | 111 ± 19a,e | 1.8 ± 0.4 | 25 ± 3.1 |
| Kidney | 148 ± 11f | 0.01 ± 0.002 | 4.6 ± 1.0b | 326 ± 49c,j | 16 ± 0.81g | 13 ± 5.3b,g,h | 51 ± 8.1i | 24 ± 1.1h | 140 ± 24f | |
| Lung | 0.41 ± 0.08 | 0.04 ± 0.01 | 5.4 ± 1.3b | 71 ± 12i | 29 ± 6.0g,j | 8.6 ± 1.9b | 45 ± 8.7i,j | 10 ± 2.7b | 8.0 ± 1.9b |
Data are the means ± SEM of mRNA molecules per pg of total RNA. Basal amounts of different Gst mRNAs from the same organ as well as basal amounts of each Gst mRNA in the three examined mouse organs were compared. Average numbers with the same superscript are not statistically different (p > 0.05).
Figure 1.
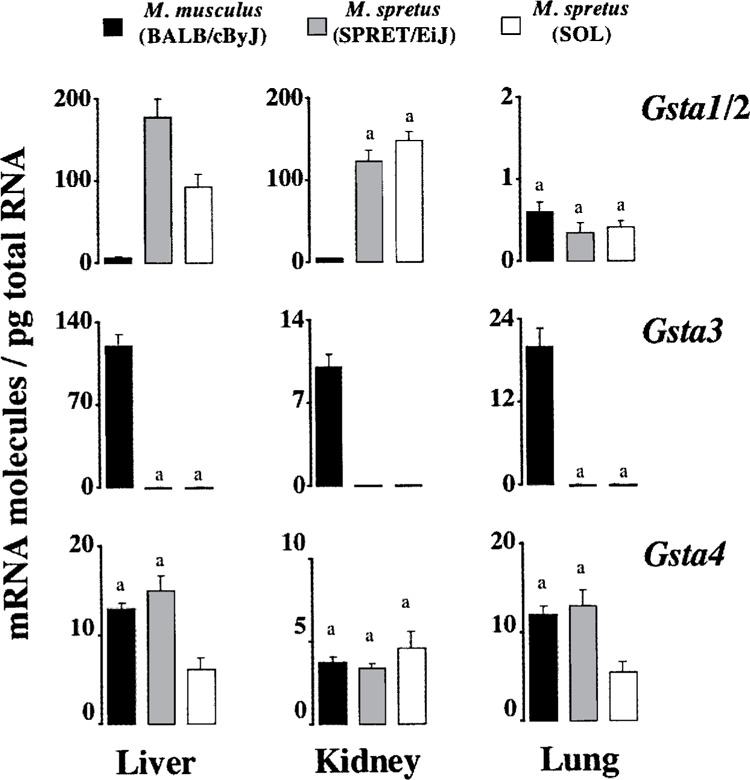
Comparison of steady-state levels of transcripts coding for alpha GSTs. Data are those reported in Tables 2, 3, and 4. Average numbers with the same superscript are not statistically different (p > 0.05).
Figure 3.
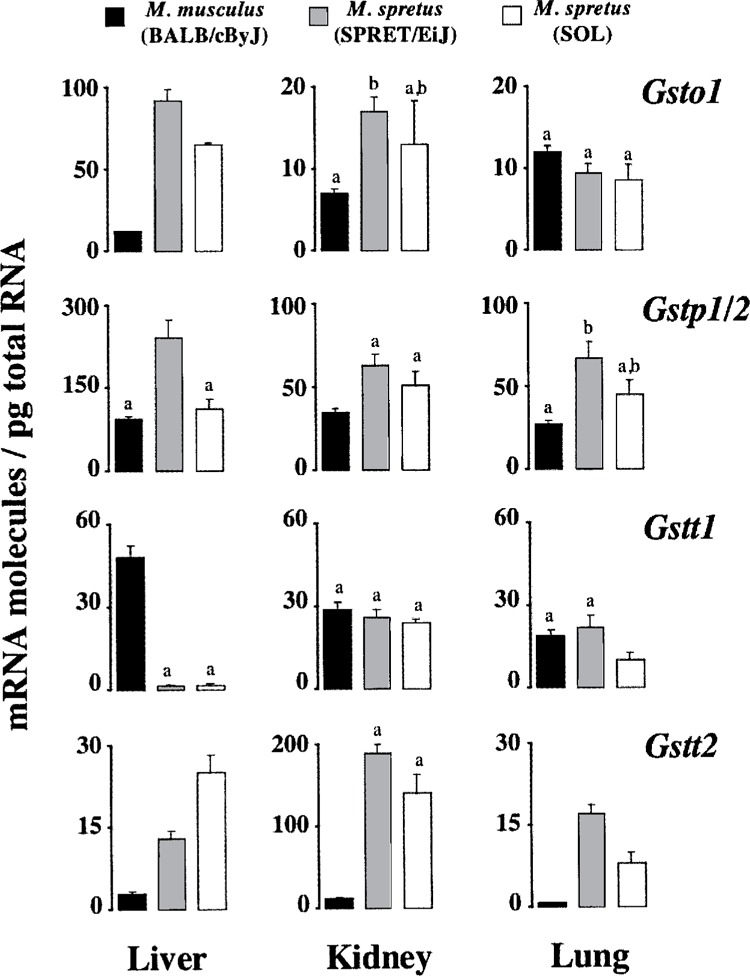
Comparison of steady-state levels of transcripts coding for omega, pi, and theta GSTs. Data are those reported in Tables 2, 3, and 4. Average numbers with the same superscript are not statistically different (p > 0.05).
Organ Expression Profiles of Basal Amounts of Gst Transcripts in M. musculus
As shown in Table 2, all transcript amounts showed extraordinary consistency between tissue samples from different animals. Overall, there was less than twofold variation in mRNA copy number between any two mice in the three studied organs. Exceptions were Gsta1/2 and particularly hepatic Gstm3 mRNA, for which MM13 displayed around fivefold greater copy number than MM09 and MM10 (9.1 vs. 1.7 and 2.0 molecules/pg).
Gstm1 was the most abundant Gst mRNA in all three organs from M. musculus, standing for 39% (lung) up to 53% (kidney) of the total Gst mRNA molecules (Table 2). Other major Gst mRNAs were Gsta3 (18%) and Gstp1/2 (14%) in liver, and Gstm2 in kidney (14%) and particularly in lung (26%). On the contrary, Gsta1/2 and Gstm3 were minor Gst mRNAs in the three M. musculus organs, where their presence does not exceed 1% of the total. Gstt2 was another mRNA of low abundance in both liver (0.4%) and lung (0.3%). Notably, in absolute terms, Gstm1 mRNA was as abundant in liver (322 molecules/pg), kidney (159 molecules/pg), and lung (100 molecules/pg) as transcripts coding for ROS-scavenging enzymes like catalase and CuZn-superoxide dismutase (data not shown), which are regarded as highly expressed proteins (16). In contrast, Gstm3 mRNA was as infrequently expressed in kidney and lung (with less than 0.1 molecules/pg) as rare splicing forms of transcript coding for cytosolic thioredoxin reductase (14).
The liver exhibited the highest levels of Gst mRNAs, followed by kidney and then lung. Hence, Gstm1, Gstp1/2, Gstt1, and particularly Gsta3 mRNAs were significantly more abundant in liver than in kidney and lung, and Gstm3 was only expressed at a considerable amount in this organ. On the contrary, Gstm1, Gstp1/2, Gstt1, Gstt2, and particularly Gsta1/2 mRNA were significantly less abundant in lung than in kidney and liver. A high steady-state level of Gstt2 mRNA, compared with liver and lung, distinguished the kidney.
Organ Expression Profiles of Basal Amounts of Gst Transcripts in M. spretus
Interindividual variation in the steady-state levels of Gst transcripts was higher in M. spretus (Tables 3 and 4) than in M. musculus (Table 2). This mouse-to-mouse variability was more pronounced among wild mice (Table 4) than among genetically homogeneous animals maintained under standard laboratory conditions (Table 3), and it depended on the Gst mRNA type and on the mouse organ. For instance, while Gsto1 had an outstanding stable expression in liver, Gsta1/2 expression in lung was quite variable among mice (Tables 3 and 4).
TABLE 3.
STEADY-STATE LEVELS OF Gst mRNAs IN M. SPRETUS (SPRET/EiJ) BRED UNDER STANDARD LABORATORY CONDITIONS
| Mouse | Gsta1/2 | Gsta3 | Gsta4 | Gstm1 | Gstm2 | Gstm3 | Gsto1 | Gstp1/2 | Gstt1 | Gstt2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Liver | 178 ± 25a,b | 0.15 ± 0.03 | 15 ± 1.7c | 638 ± 51 | 239 ± 39a | 0.02 ± 0.003 | 92 ± 7.0 | 242 ± 32a | 1.8 ± 0.23 | 13 ± 1.3c |
| Kidney | 123 ± 14b | 0.03 ± 0.004d | 3.4 ± 0.24 | 418 ± 30 | 17 ± 3.3e,f | 17 ± 1.8e | 63 ± 6.8g | 26 ± 2.8f | 189 ± 11 | |
| Lung | 0.35 ± 0.12 | 0.04 ± 0.005d | 13 ± 1.8c,h,i | 96 ± 12j | 48 ± 6.3g | 0.001 ± 0.0002 | 9.4 ± 1.2h | 67 ± 10g,j | 22 ± 4.1f,i | 17 ± 1.7c,i |
Data are the means ± SEM of mRNA molecules per pg of total RNA. Basal amounts of different Gst mRNAs from the same organ as well as basal amounts of each Gst mRNA in the three examined mouse organs were compared. Average numbers with the same superscript are not statistically different (p > 0.05).
Like in M. musculus (Table 2), Gstm1 was the most abundant of the investigated transcripts in all three organs from M. spretus, comprising from approx. 37% (lung) to 47% (kidney) of total Gst mRNA molecules (Tables 3 and 4). In M. spretus, however, other major Gst mRNAs were Gstm1 and Gstp1/2 in both liver and lung, Gsta1/2 in both liver and kidney, and Gstt2 in kidney. On the other hand, Gsta3 and particularly Gstm3 were very rare transcripts in all three organs examined, where their amounts range from ∼0.1 (liver Gsta3) to ≤0.001 molecules/pg (kidney and lung Gstm3). Gsta1/2 and Gstt1 were other mRNAs of low abundance in lung and liver, respectively.
Species-Associated Differences in Basal Gst mRNA Levels
The steady-state Gst mRNA levels in M. musculus and M. spretus are compared in Figures 1–3, to make clear that the expression profiles obtained are remarkably different in an absolute quantitative sense. Of particular interest were the pronounced differences seen with transcripts coding for GSTs belonging to the class alpha (Fig. 1). The Gsta3 mRNA levels in M. musculus exceed greatly those quantitated in M. spretus. In liver, for instance, that means a drop from 120 molecules/pg to just about 0.1 molecules/pg. Remarkably, this Gsta3 downregulation went along with the upregulation of Gsta1/2 mRNA levels in liver and also kidney of M. spretus. By means of the primers and the PCR amplification conditions described previously (5), we further established (data not shown) that, although the Gsta2 mRNA is the predominant transcript (accounting for >70% of total Gsta1 and Gsta2 molecules), both Gsta1 and Gsta2 mRNAs were increased in liver and kidney (not lung) of M. spretus. For instance, basal amounts of renal Gsta1 and Gsta2 mRNAs were determined to be 11 and 125 molecules/pg, respectively, in M. spretus (SPRET/EiJ) but 0.2 and 1 molecules/pg in M. musculus. In contrast to Gsta1/2, no compensatory changes were clearly appreciated in basal Gsta4 expression (Fig. 1).
With respect to transcripts coding for GSTs of the class theta (Fig. 3), Gstt1 transcripts from liver were less abundant in M. spretus (1.8 molecules/pg) than in M. musculus (48 molecules/pg), this decrease being accompanied by an increment in Gstt2 mRNA molecules. Such an increase in Gstt2 transcript levels was also observed in kidney and lung, though in these two organs Gstt1 mRNA showed a remarkably consistent level of expression in the two mouse species. Interestingly, in kidney from M. spretus Gstt2 mRNA reached a copy number relatively close to that of the most abundant Gstm1 transcript (Fig. 2), and similar to that of Gsta1/2 mRNAs (Fig. 1). Other interesting observations were the absence or extremely low expression of Gstm3 in M. spretus (Fig. 2), and the upregulation of the most recently discovered Gsto1 (3,17) in liver from this mouse species (Fig. 3). Overall, total Gst mRNA molecules quantitated herein were higher in liver and kidney from M. spretus than from M. musculus, the lung being the organ with lowest expression levels and fewest differences between both species.
Figure 2.
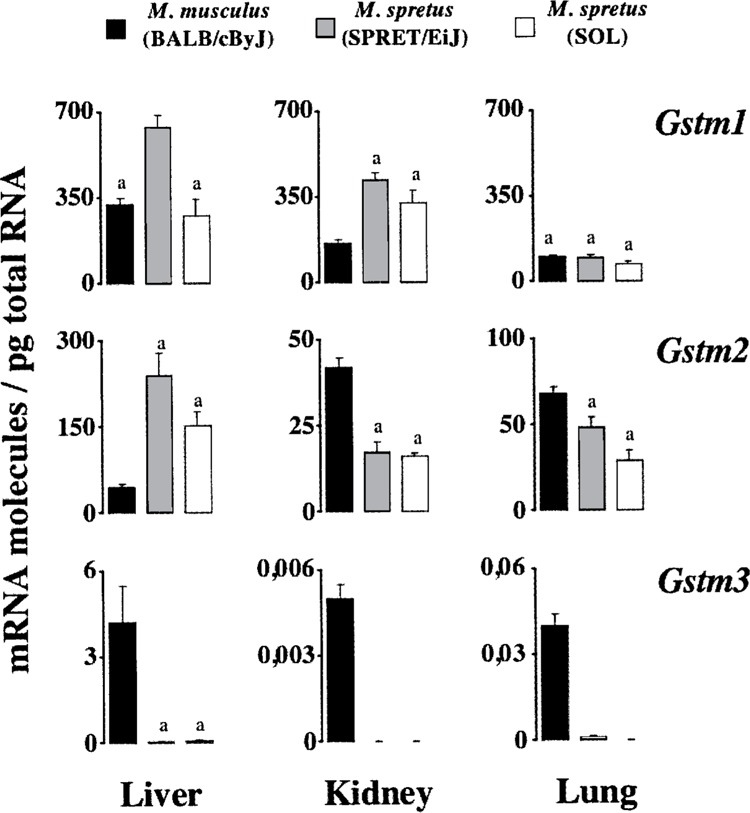
Comparison of steady-state levels of transcripts coding for mu GSTs. Data are those reported in Tables 2, 3, and 4. Average numbers with the same superscript are not statistically different (p > 0.05).
In contrast to the outstanding differences found between M. musculus and M. spretus, the Gst mRNA levels in wild M. spretus were in most cases (19/30) nonstatistically different to those quantitated in wild-derived inbred animals (Figs. 1–3). On the whole, major differences were observed in the liver and minor differences in the kidney, where only the extremely low copy numbers of Gsta3 mRNA were found significant between both mouse groups (0.01 vs. 0.03 molecules/pg). Interestingly, in general, wild animals from the nonpolluted SOL sampling area exhibited lower Gst mRNA levels than wild-derived inbred mice.
Gst mRNA Levels Following Paraquat Exposure
Protection against oxidative stress is an ancient but ongoing problem for living organisms. To quantitate the abundance of Gst transcripts under an oxidizing condition, M. musculus male mice were challenged with the herbicide PQ (a superoxide generator) through both time course and dosage–response experiments. The liver is often the target organ of toxic compounds, but the lung is considered the primary target organ of PQ toxicity [a well-known pneumotoxicant; e.g., see (2,24)]. Hence, to shorten the analysis, quantitations were limited to lungs from both challenged and control mice. Control animals (saline IP) showed Gst transcript levels (Fig. 4) similar to those quantified in noninjected animals (Table 2). In addition, no time- or dosage-related effect was noted in these vehicle controls (data not shown), in contrast to the PQ-challenged animals.
Figure 4.
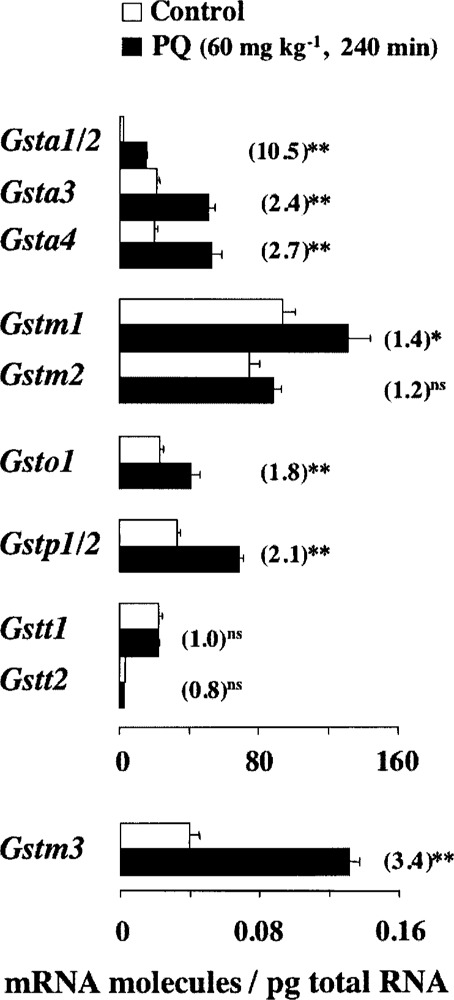
Pulmonary response to oxidative stress in M. musculus. Data are the means of mRNA molecules/pg of total RNA ± SEM from 12 control and 3 challenged mice, respectively. To facilitate the comparison between relative and absolute quantitations, data from PQ-treated animals were divided by those of control animals. These fold increments are given in parentheses. Statistical significance is expressed as: **p < 0.01, *p < 0.05, and ns p > 0.05.
Quantitations of pulmonary transcripts revealed that the steady-state amounts of Gsta1/2, Gsta3, Gsta4, Gstm3, Gsto1, and Gstp1/2 are readily upregulated by PQ in a time- and/or dose-dependent manner (data not shown). Concerning the expression of Gstm1 gene, the only effect toward significance was observed at 60 mg kg−1 body weight for 240 min of challenge, the PQ threatening condition that also gave maximal induction levels for the rest of the upregulated Gst mRNAs (Fig. 4). In contrast, no significant induction of Gstm2, Gstt1, and Gstt2 mRNA levels was detected in lung through any of the PQ exposure conditions investigated herein. In fact, the Gstm2 and Gstt1 mRNA levels were moderately downregulated within the first hour of PQ exposure; over time those transcript levels were gradually adjusted to the amounts seen in unstressed animals (Fig. 4 and data not shown).
The conventional fold variations calculated in Figure 4 highlight the meaning of the absolute measurements reported in the current study. For instance, although a relative increment of about 2.5-fold in Gsta3 and Gsta4 transcript levels might look modest compared to the 10.5-fold increase in Gsta1/2 mRNA, the actual scenario is that, after PQ exposure, Gsta3 and Gsta4 mRNAs are still present in higher amounts than Gsta1/2 mRNA (52 vs. 15 molecules/pg). Likewise, after the 3.4-fold induction of pulmonary message for Gstm3, this transcript remains at much lower amount than those for Gstm1 or Gstm2 (0.13 vs. 132 and 88 molecules/pg, respectively).
Sampling Area-Associated Differences in Gst mRNA Levels
Upon establishing the response to PQ, we determined whether the Gst mRNA levels would discriminate between M. spretus mice dwelling in sites with low or high environmental pollution. The reference site was the Santa Olalla Lagoon (SOL) within the Doñana Biological Reserve. The polluted site (PS) was located in an industrial settlement consisting of a large petrochemical complex, several phosphoric acid plants specialized in the production of fertilizers from phosphorites, and some chalcopyrite transformation plants (7). The polluted site is about 50 km NW the reference.
Ten male mice of 11 ± 0.5 g body weights were captured in each sampling site. To shorten the analysis, the Gst transcript levels were quantitated in pooled samples of total RNA from individual lungs of mice from each site. No differences were found between sites for most Gst mRNAs. The exception was the fivefold increase (1.9 vs. 0.38 molecules/pg) in Gsta3 mRNA copy number that was quantitated in mice from PS compared to those from SOL. Unexpectedly, however, the 0.38 Gsta3 mRNA molecules/pg measured in the pooled RNA sample from SOL animals was clearly higher than those reported in Table 4 (0.04 ± 0.01 molecules/pg). To gain further insight into the nature of such a difference, Gsta3 transcript levels were quantified on individual lungs from mice of the two sampling areas. As shown in Figure 5, 1 out of the 10 mice captured at SOL showed an outstanding high level of Gsta3 mRNA (3.4 vs. the average value of 0.05 molecules/pg). Interestingly, however, the samples from the PS polluted location showed higher variability in Gsta3 mRNA copy number than those from the reference site, with 3 of the 10 lungs analyzed exhibiting >50-fold higher amounts than average. No statistical significance between mice collected in the two sites was reached due to such an interindividual variation.
Figure 5.
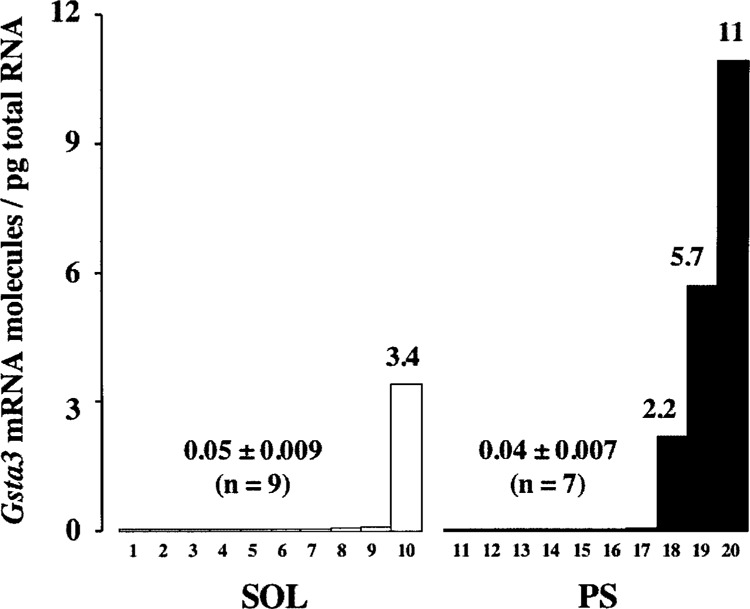
Pulmonary Gsta3 mRNA levels in M. spretus collected in the SOL and PS sites. Each bar represents the number of Gsta3 mRNA molecules/pg of total RNA in the lung from a single mouse. In the x-axis, lungs are numbered consecutively from lowest to greatest amounts of Gsta3 transcript level.
Interindividual differences may be attributable to genetic variation in the population, or they may be due to other factors, such as animal diet, sex and age, and habitat contamination [e.g., see (8)]. In this line, the increased Gsta3 mRNA level detected in 1 of the 10 mice collected at the reference site may be an indication of the progressive pollution of key Doñana ecosystems, as reported by other studies (4).
DISCUSSION
Differential expression of mammalian genes is critical for both normal cellular processes and abnormal processes associated with disease. Investigations into the transcriptional expression of murine Gst genes have been restricted to inbred strains for laboratory use, and they have used analytical approaches that generate at best semiquantitative/relative data [e.g. (5)]. The purpose of the present work was to make use of a refined and accurate RT-PCR strategy (14) to provide the first comprehensive and absolute quantitative analysis of the overall expression patterns of transcripts coding for most murine GSTs of alpha, mu, omega, pi, and theta families. In line with this end, quantitations were performed to examine how the steady-state levels of Gst transcripts are modulated in association with: three animal organs where extensive detoxification reactions occur (liver, kidney, and lung), two mouse species representing common laboratory and bioindicator animals (M. musculus and M. spretus), and two distinct genetic and animal living conditions (wild-derived inbred animals vs. free-living mice). Moreover, quantitations performed examined how those steady-state Gst mRNA amounts are affected in M. musculus by the oxidizing conditions posed by PQ (a potent herbicide and human poison), and in M. spretus dwelling at a polluted area.
The absolute quantitations of the steady-state levels of Gst transcript molecules revealed both organ-specific (Tables 2–4) and species-specific (Figs. 1–3) patterns of expression. Whereas the tissue-specific expression of GSTs of classes alpha, mu, omega, pi, and theta has been examined previously [see, e.g., (9,10,17,26)], little work has been done on the comparison of Gst mRNA expression in different species of mouse. Concerning tissue differences, published data (obtained typically by in situ hybridization, multiple-tissue Northern blot, and/or RT-PCR) are usually imprecise in quantitative terms, and not in general agreement with the absolute measurements given in the present study. For instance, in contrast to the results in Table 2: i) image analysis of in situ hybridization data did not reveal differences among relative quantities of Gsta1, Gstm1, and Gstp1 mRNAs in murine lung (10); ii) Northern blot analysis of total RNA from a variety of mouse tissues showed that whereas both Gstt1 and Gstt2 transcripts are abundantly expressed in liver and kidney, their expression is barely detectable in lung (26); and iii) quantitative real-time PCR data indicated that the relative content of hepatic Gsta4 mRNA is far lower than those of Gsta1/2 and Gsta3 mRNAs (9- and 32-fold, respectively) (9).
Concerning the mouse species (Figs. 1–3 and data not shown), we quantitated for the first time outstanding differences in the steady-state amounts of individual Gst mRNAs (i.e., Gsta1, Gsta2, Gsta3, Gstm2, Gstm3, Gsto1, Gstt1, and Gstt2). These differences might have important physiological consequences in terms of animal susceptibility to the cytotoxic and genotoxic effects of xenobiotics. In view of the reported catalytic properties of GSTs in the mouse, some predictions are possible [see, e.g., (5)]. For instance, liver and kidney of M. spretus mice that express Gsta1/2 mRNAs at an increased level should be more resistant to polycyclic aromatic hydrocarbons than M. musculus mice. In contrast, M. spretus mice should be intrinsically at high risk to the hepatocarcinogen aflatoxin B1, considering the about 1000-fold less hepatic Gsta3 mRNA copies quantitated in this species compared with M. musculus. On the other hand, the ability of mammalian Gstt1 to activate dihaloalkanes into mutagenic half-mustards [reviewed in (12,18)] anticipates that the low Gstt1 mRNA copy number quantitated in liver from M. spretus will confer to this mouse species special resistance to the genotoxic action of such an important group of common environmental pollutants.
The murine Gsta1 gene contains in its promoter a tandem repeat of the antioxidant-responsive element (ARE) (11). A single A→G transition that confers increased expression has been detected within the ARE motif of the Gsta1 promoter in M. spretus (liver from inbred females), compared with M. musculus (BALB/cByJ) (27). This variation might explain the enhanced steady-state Gsta1 mRNA levels that we quantified in both liver and kidney (not lung) of male M. spretus (inbred or free-living animals). In this line, the large differences reported here in the steady-state amounts of most investigated Gst mRNAs (besides that of Gsta1) between M. spretus and M. musculus encourage further studies on additional naturally occurring polymorphisms in cis-acting regulatory elements. This knowledge will help for a better understanding of regulation of Gst gene transcription (e.g., with the exception of Gsta1, no other mouse Gst genes are known to contain functional AREs), and in determining cancer risk in human and other mammals.
Previous studies (14,19) from this laboratory have shown that the copy number of various mRNAs coding for proteins with antioxidative activity are upregulated in vivo in response to the superoxide stress posed by the widely used herbicide PQ. These transcripts code for major AP-endonuclease (AP-1), heme oxygenase (HO-1), and cytosolic thioredoxin reductase (TrxR1), and for the single glutathione reductase (GR). In this work we demonstrate that, in addition to these mRNAs, the amounts of transcripts coding for GSTs are also responsive to PQ (Fig. 4), in agreement with the central role attributed to these enzymes in defense against oxidative stress.
Given that, to the best of our knowledge, we provided here the first in vivo evidence that pulmonary Gst mRNA levels are influenced by PQ treatments, no detailed comparisons with previous works are possible. Nonetheless, our results as a whole are in line with earlier studies demonstrating that the levels of transcripts coding for GSTs belonging to the alpha, mu, and pi families are increased under oxidizing conditions [see, e.g., (5,8)]. A striking additional observation of the present study is the significant induction of the most recently discovered Gsto1 gene (3,17), unlike those coding for the theta class GSTs. From a quantitative perspective, it is also of note the finding that among the PQ-responsive genes coding for GSTs of the same class, those expressed at high abundance constitutively exhibited the greatest increases in absolute copy numbers, though the greatest relative fold increases were observed for those mRNAs of low steady-state levels. Hence, we propose that the establishment of irrefutable methods for quantitation of actual mRNA copy numbers, as described here, should provide a basis for interpreting and planning future studies on transcriptional regulation of murine Gst genes (e.g., to investigate the apparent dissociation between the involvement of Nrf2 regulator in basal and oxidative inducible expression) [see, e.g., (5)].
Whereas the PQ treatments aimed to induce in animals an acute oxidative stress response, the field study was designed to evaluate the long-term effects on Gst gene expression of an environmental chronic stress. Although preliminary, data reported suggest that M. spretus mice dwelling in one of the largest protected area in Europe (Doñana) have lower and more homogeneous mRNA levels of the Gsta3 gene than those collected in anthropogenic environments, such as industrial settlements around petrochemical plants. This preliminary finding encourages further studies, both by extending the analyses to other murine organs and wild populations of both sexes, and particularly by studying (as discussed above) the molecular mechanism controlling both basal and induced Gst mRNA levels. In any case, our study suggests that absolute quantitation of transcripts in free-living organisms can be a sensitive end-point as indicator of environmental stress, in addition to other more conventional genetic and biochemical analysis.
ACKNOWLEDGMENT
This work was supported by Ministerio de Ciencia y Tecnología (grants BMC2002-00179 and REN2002-04366-CO2-01).
REFERENCES
- 1. Adler D. A.; Rugarli E. I.; Lingenfelter P. A.; Tsuchiya K.; Poslinski D.; Liggitt H. D.; Chapman V. M.; Elliott R. W.; Ballabio A.; Disteche C. M. Evidence of evolutionary up-regulation of the single active X chromosome in mammals based on Clc4 expression levels in Mus spretus and Mus musculus . Proc. Natl. Acad. Sci. USA 94:9244–9248; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ali S.; Diwakar G.; Pawa S. Paraquat induces different pulmonary biochemical responses in Wistar rats and Swiss mice. Chem. Biol. Interact. 125:79–91; 2000. [DOI] [PubMed] [Google Scholar]
- 3. Board P. G.; Coggan M.; Chelvanayagam G.; Easteal S.; Jermiin L. S.; Schulte G. K.; Danley D. E.; Hoth L. R.; Griffor M. C.; Kamath A. V.; Rosner M. H.; Chrunyk B. A.; Perregaux D. E.; Gabel C. A.; Geoghegan K. F.; Pandit J. Identification, characterization, and crystal structure of the omega class glutathione transferases. J. Biol. Chem. 275:24798–24806; 2000. [DOI] [PubMed] [Google Scholar]
- 4. Bonilla-Valverde D.; Ruiz-Laguna J.; Muñoz A.; Ballesteros J.; Lorenzo F.; Gómez-Ariza J. L.; López-Barea J. Evolution of biological effects of Aznal-collar mining spill in the Algerian mouse (Mus spretus) using biochemical biomarkers. Toxicology 197:123–138; 2004. [DOI] [PubMed] [Google Scholar]
- 5. Chanas S. A.; Jiang Q.; McMahon M.; McWalter G. K.; McLellan L. I.; Elcombe C. R.; Henderson C. J.; Wolf C. R.; Moffat G. J.; Itoh K.; Yamamoto M.; Hayes J. D. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 365:405–416; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coviello-McLaughlin G. M.; Prowse K. R. Telomere length regulation during postnatal development and ageing in Mus spretus . Nucleic Acids Res. 25:3051–3058; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Degrassi F.; Tanzarella C.; Ieradi L. A.; Zima J.; Cappai A.; Lascialfari A.; Allegra F.; Cristaldi M. CREST staining of micronuclei from free-living rodents to detect environmental contamination in situ . Mutagenesis 14:391–396; 1999. [DOI] [PubMed] [Google Scholar]
- 8. Edwards M. G.; Sarkar D.; Klopp R.; Morrow J. D.; Weindruch R.; Prolla T. A. Age-related impairment of the transcriptional responses to oxidative stress in the mouse heart. Physiol. Genomics 13:119–127; 2003. [DOI] [PubMed] [Google Scholar]
- 9. Engle M. R.; Singh S. P.; Czernik P. J.; Gaddy D.; Montague D. C.; Ceci J. D.; Yang Y.; Awasthi S.; Awasthi Y. C.; Zimniak P. Physiological role of mGSTA4-4, a glutathione S-transferase metabolizing 4-hydroxynonenal: Generation and analysis of mGsta4 null mouse. Toxicol. Appl. Pharmacol. 194:296–308; 2004. [DOI] [PubMed] [Google Scholar]
- 10. Forkert P. G.; D’Costa D.; El-Mestrah M. Expression and inducibility of alpha, pi, and mu glutathione S-transferase protein and mRNA in murine lung. Am. J. Respir. Cell. Mol. Biol. 20:143–152; 1999. [DOI] [PubMed] [Google Scholar]
- 11. Friling R. S.; Bensimon A.; Tichauer Y.; Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. USA 87:6258–6262; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayes J. D.; Strange R. C. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology 61:154–166; 2000. [DOI] [PubMed] [Google Scholar]
- 13. Hochepied T.; Schoonjans L.; Staelens J.; Kreemers V.; Danloy S.; Puimege L.; Collen D.; Van Roy F.; Libert C. Breaking the species barrier: Derivation of germline-competent embryonic stem cells from Mus spretus × C57BL/6 hybrids. Stem Cells 22:441–447; 2004. [DOI] [PubMed] [Google Scholar]
- 14. Jurado J.; Prieto-Álamo M. J.; Madrid-Rísquez J.; Pueyo C. Absolute gene expression patterns of thioredoxin and glutaredoxin redox systems in mouse. J. Biol. Chem. 278:45546–45554; 2003. [DOI] [PubMed] [Google Scholar]
- 15. Ketterer B. A bird’s eye view of the glutathione transferase field. Chem. Biol. Interact. 138:27–42; 2001. [DOI] [PubMed] [Google Scholar]
- 16. Kitteringham N. R.; Powell H.; Jenkins R. E.; Hamlett J.; Lovatt C.; Elsby R.; Henderson C. J.; Wolf C. R.; Pennington S. R.; Park B. K. Protein expression profiling of glutathione S-transferase pi null mice as a strategy to identify potential markers of resistance to paracetamol-induced toxicity in the liver. Proteomics 3:191–207; 2003. [DOI] [PubMed] [Google Scholar]
- 17. Kodym R.; Calkins P.; Story M. The cloning and characterization of a new stress response protein. A mammalian member of a family of theta class glutathione s-transferase-like proteins. J. Biol. Chem. 274:5131–5137; 1999. [DOI] [PubMed] [Google Scholar]
- 18. Landi S. Mammalian class theta GST and differential susceptibility to carcinogens: A review. Mutat. Res. 463:247–283; 2000. [DOI] [PubMed] [Google Scholar]
- 19. Prieto-Álamo M. J.; Cabrera-Luque J. M.; Pueyo C. Absolute quantitation of normal and ROS-induced patterns of gene expression: An in vivo real-time PCR study in mice. Gene Expr. 11:23–34; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pueyo C.; Jurado J.; Prieto-Álamo M. J.; Monje-Casas F.; López-Barea J. Multiplex reverse transcription-polymerase chain reaction for determining transcriptional regulation of thioredoxin and glutaredoxin pathways. Methods Enzymol. 347:441–451; 2002. [DOI] [PubMed] [Google Scholar]
- 21. Ruíz-Laguna J.; García-Alfonso C.; Peinado J.; Moreno S.; Ieradi L. A.; Cristaldi M.; López-Barea J. Biochemical biomarkers of pollution in Algerian mouse (Mus spretus) to assess the effects of the Aznal-cóllar disaster on Doñana Park (Spain). Biomarkers 6:146–160; 2001. [DOI] [PubMed] [Google Scholar]
- 22. Salinas A. E.; Wong M. G. Glutathione S-transferases-a review. Curr. Med. Chem. 6:279–309; 1999. [PubMed] [Google Scholar]
- 23. Totsuka Y.; Nagao Y.; Horii T.; Yonekawa H.; Imai H.; Hatta H.; Izaike Y.; Tokunaga T.; Atomi Y. Physical performance and soleus muscle fiber composition in wild-derived and laboratory inbred mouse strains. J. Appl. Physiol. 95:720–727; 2003. [DOI] [PubMed] [Google Scholar]
- 24. Venkatesan N. Pulmonary protective effects of curcumin against paraquat toxicity. Life Sci. 66:PL21–28; 2000. [DOI] [PubMed] [Google Scholar]
- 25. Viegas-Crespo A. M.; Lopes P. A.; Pinheiro M. T.; Santos M. C.; Rodrigues P. D.; Nunes A. C.; Marques C.; Mathias M. L. Hepatic elemental contents and antioxidant enzyme activities in Algerian mice (Mus spretus) inhabiting a mine area in central Portugal. Sci. Total Environ. 311:101–109; 2003. [DOI] [PubMed] [Google Scholar]
- 26. Whittington A.; Vichai V.; Webb G.; Baker R.; Pearson W.; Board P. Gene structure, expression and chromosomal localization of murine theta class glutathione transferase mGSTT1-1. Biochem. J. 337 (Pt. 1):141–151; 1999. [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu M.; Chapman W. G.; Oberley M. J.; Wasserman W. W.; Fahl W. E. Polymorphic electrophile response elements in the mouse glutathione S-transferase GSTa1 gene that confer increased induction. Cancer Lett. 164:113–118; 2001. [DOI] [PubMed] [Google Scholar]


