Abstract
Broad differentiation capacity has been described for mesenchymal stem cells (MSC) from human bone marrow. We sought to identify genes associated with the immature state and pluripotency of this cell type. To prove the pluripotent state of the MSC, differentiation into osteocytes, adipocytes, and chondrocytes was performed in vitro. In contrast, normal skin cells did not harbor these differentiation abilities. We compared the expression profile of human bone marrow MSC with cDNA from one primary human skin cell line as control, using a cDNA chip providing 9600 genes. The identity of all relevant genes was confirmed by direct sequencing. Data of gene array expression were corroborated employing quantitative PCR analysis. About 80 genes were differently expressed more than threefold in MSC compared to mature skin fibroblasts. Interestingly, primary human MSC were found to upregulate a number of genes important for embryogenesis such as distal-less homeo box 5, Eyes absent homolog 2, inhibitor of DNA binding 3, and LIM protein. In contrast, mesenchymal lineage genes were downregulated in MSC in comparison to skin cells. We also detected expression of some genes involved in neural development, indicating the broad differentiation capabilities of MSC. We conclude that human mesenchymal stem cells harbor an expression profile distinct from mature skin fibroblast, and genes associated with developmental processes and stem cell function are highly expressed in adult mesenchymal stem cells.
Key words: Mesenchymal stem cells, Microarray, Differentiation, Gene expression profile
MESENCHYMAL stem cells (MSC) from human bone marrow are one of the most accessible adult stem cells described to date. Mesenchymal stem cells have first been described as fibroblasts colony-forming cells (24,25). Due to their differentiation abilities they recently have been termed marrow stromal cells (69), mesodermal progenitor cells (MPC) (73), or multipotent adult progenitor cells (MAPC) (31). MSC can be differentiated in vitro into osteocytes, chondrocytes, adipocytes, myocytes, endothelial cells, and hematopoesis-supporting stroma (68,73). They seem to contribute to most, if not all, somatic cell types, when injected into early blastocysts or into xenogenic embryos (31,43). In contrast, skin fibroblasts, the mature counterpart of MSC, do not harbor these broad differentiation abilities (68). Yet, little is known about genes and signal transduction pathways involved in the maintenance of pluripotency or its loss.
Microarray analysis is a useful screening technique for gene expression profiles. Therefore, we compared the expression profile of MSC cDNA obtained from human bone marrow of four healthy donors with that from primary human skin fibroblasts derived from healthy donors as control. Employing this technique we were interested to investigate gene expression profiles of MSC that could be associated with the pluripotent state and differentiation capacity of MSC.
MATERIALS AND METHODS
Mesenchymal Stem Cell Culture
The culture methods of Pittenger (68) were employed here with minor modifications. Spongiform bone fragments from hip replacement operations were taken from otherwise healthy volunteers after written informed consent at one institution (Philipps-University, Marburg). The protocol had been approved by our local Ethics Committee. Cells were processed within 12 h after collection. Specimens were thoroughly minced, passed through a 70-μm filter mesh, and subsequently mononuclear cells (MNCs) were obtained by Ficoll™ (Amersham Pharmacia, NJ, USA) density gradient centrifugation, washed with phosphate-buffered saline (PBS) (Biochrom, Berlin, Germany), and resuspended in Dulbecco’s modified medium (DMEM) with low glucose and glutamine (PAA, Linz, Austria) with 10% fetal calf serum (FCS) from selected lots (Stem Cell Technologies, Vancouver, Canada) and 1% penicillin/streptomycin. Cells were plated at a concentration of 200,000 cell/cm2 in plastic flasks. Medium was replaced after the first 24 h and then every 3–4 days until fibroblasts grew confluent. Cells were then collected by treatment with trypsin/EDTA solution (Roche, Basel, Switzerland) for 10 min at 37°C, rinsed with medium, and split for three new culture dishes of the same size. With every splitting and exchange of a culture dish the passage number was increased.
To ensure purity of MSC, flasks were washed two times thoroughly with PBS and the medium was replaced within 24 h. With this technique contamination of hematopoietic stem cells, macrophages, or endothelial cells could be avoided effectively as proved by flow cytometric antigen expression analysis. For staining of surface molecules, 105 to 106 cells were incubated in 100 μl PBS with 10 μl of fluorochrome-labeled monoclonal antibody for 20 min at 4°C in darkness. Then cells were washed with PBS, resuspended in 300 μl PBS, and immediately submitted to FACS analysis (FACScan, Becton Dickinson, San Jose, CA, USA). For the CD105 antibody (PharMingen, San Jose, CA, USA) indirect staining was performed with incubation of the primary antibody for 20 min at 4°C, two times washing, staining for 10 min with the goat anti-mouse PE secondary antibody (PharMingen), washing with PBS, and resuspension in 300 μl PBS for further analysis. The following antibodies were employed: CD45-FITC (PharMingen), CD90-FITC (PharMingen), CD34-PE (PharMingen), CD14-PE (Becton Dickinson), CD31-PE (Beckmann-Coulter, Krefeld, Germany), CD44-FITC (Becton Dickinson), IgG-FITC and IgG-PE (Becton Dickinson).
Cloning efficiency of MSC had been investigated under different conditions (different FCS lots, different media, use of conditioned media). MSC culture could be maintained for more than 20 passages (1 year) and efficient cloning was proved by CFU-fibroblast (CFU-F) formation even after 7 months of in vitro culture. We used MSC cells before the third passage for microarray experiments and for further differentiation procedures.
Cell Cultures of Skin Fibroblasts
Small pieces of skin from the patients undergoing hip replacement were cut into tiny pieces and then digested with 0.2% collagenase (Roche, Basel, Switzerland) for 15 min at 37°C, followed by 0.1% trypsin incubation (Invitrogen, Grand Island, NY, USA) for 30 min at 37°C. The tissue was triturated briefly and then passed through a 70-μm filter. Cells were collected by centrifugation and then resuspended in DMEM medium with 10% FCS (Gibco BRL, Grand Island, NY, USA) and 1% penicillin/streptomycin. Medium was replaced every 3–4 days until the cells grew confluent. Skin fibroblasts were harvested by treating with trypsin/EDTA solution and were split into three portions for three new culture flasks. For microarray experiments and differentiation skin fibroblasts were harvested at the second passage.
Osteogenic Differentiation
For osteogenic differentiation the cells were harvested as described above and were allowed to adhere within 24 h in standard medium in humidified air of 5% CO2 at 37°C. Subsequently, standard medium was replaced through osteogenesis induction medium with 10% FCS from selected lots (Stem Cell Technologies), DMEM low glucose with glutamine, 0.1 μM dexamethasone, 0.05 mM ascorbic acid-2-phosphate, and 10 mM β-glycerophosphate (all obtained from Sigma, Taufkirchen, Germany). Medium was replaced every 3–4 days. After 10–14 days of culture, cells were harvested for FACS analysis as described below. Slides were fixed with acetone/methanol (1:1) at −20°C for 5 min to perform hematoxylin & eosin (H&E) and von Kossa staining of the extracellular calcium matrix as described (Poetics/Bio Whittaker/Cambrex; Instructions for use of mesenchymal stem cells) (68).
Chondrogenic Differentiation
To induce chondrogenic differentiation 2.5 × 105 MSC or skin fibroblasts were cultured in 15 ml conical polypropylene tube chondrogenesis induction medium consisting of the following ingredients: DMEM high-glucose medium, 0.1 μM dexamethasone, 1 mM sodium pyruvate, 0.17 mM ascorbic acid-2-phosphate, 0.35 mM proline, 6.25 μg/ml bovine insulin, 6.25 μg/ml transferrin, 6.25 μg/ml selenous acid, 5.33 μg/ml linoleic acid, 1.25 μg/ml BSA, and 0.01 μg/ml TGF-β3 (all obtained from Stem Cell Technologies). Cells were cultured in humidified air of 5% CO2 at 37°C with a medium change every 3–4 days and were harvested after 21 days in culture. Only MSC after induction of differentiation but not the skin fibroblast or the control cells, which were pelleted in the same way but cultured in standard medium, formed small solid aggregates due to generation of surrounding solid matrix. The small cartilage piece and the loose aggregates from skin fibroblast differentiation and controls were embedded in paraffin for microdissections and subsequently stained with Alzian blue and nuclear fast red stain to characterize the proteoglycan extracellular matrix and nuclei as described.
Adipogenic Differentiation
For adipogenic differentiation the cells were cultured in standard medium. After confluence they grew 3–4 days in standard medium and subsequently the medium was replaced through adipogenic induction medium for 3 days. The adipogenic induction medium consisted of DMEM high-glucose medium (PAA), 10% FCS (Stem Cell Technologies), 1% penicillin/streptomycin, 1 μM dexamethasone, 0.2 mM indomethacin, 0.01 mg/ml insulin, 0.5 mM 3-isobutyl-1-methyl-xanthine (Sigma, Taufkirchen, Germany). After 3 days the adipogenic induction medium was replaced through adipogenic maintenance medium, consisting of DMEM high-glucose medium, 10% FCS from selected lots, 1% penicillin/streptomycin, 0.01 mg/ml insulin. This treatment of medium replacement was repeated three times. The cultures were maintained in adipogenic maintenance medium for 1 week. After this time the slides were fixed and stained with H&E and Sudan black B to visualize the lipid vacuoles. The cells were harvested, RNA was extracted, and RT-PCR was performed to analyze the expression of peroxisome proliferator-activated receptor-γ2 (PPARγ2) as described.
Flow Cytometric Analyses of Differentiation
The cells were harvested with trypsin/EDTA, washed by centrifugation with PBS, and 105 to 106 cells were incubated in 100 μl PBS with 10 μl fluorochrome-labeled monoclonal antibody for 20 min at 4°C in the dark. Subsequently cells were washed and resuspended in 500 μl PBS and FACS analysis was immediately performed using a FACScan analyser (Becton Dickinson). For the anti-bone sialoprotein antibody (Calbiochem, San Diego, CA, USA) indirect staining was performed with 10 μl primary antibody. After incubation for 20 min at 4°C and two times washing with PBS labeled cells were stained with secondary FITC goat anti-mouse antibody (PharMingen) for 10 min. They were washed two times in PBS and analyzed immediately. The following antibodies were used: CD45-FITC (PharMingen), CD90-FITC (PharMingen), CD34-PE (PharMingen), CD14-FITC (Becton Dickinson), CD44-FITC (Becton Dickinson), CD105-PE (CALTAG, Burlingame, CA, USA), anti-FITC-alkaline phosphatase (LAP) antibody (Pharmigen), anti-bone sialoprotein (BSP) antibody (Calbiochem), and IgG-FITC and IgG-PE (Becton Dickinson). The employed LAP antibody clone has been reported to recognize human bone alkaline phosphatase (51,56). Bone sialoprotein is known as initiator of matrix mineralization and is upregulated during the mineralization phase of osteoblast differentiation (27).
cDNA Microarray
We employed a custom-made cDNA microarry chip that was constructed at the Institute for Molecular Biology and Tumor Research Marburg as described previously (8). There were 9600 independent cDNA clones selected from a cDNA library obtained from Research Genetics (Invitrogen Corporation, Carlsbad, CA, USA). RNA was amplified in vitro according to modified “antisense” method (5,66). cDNA synthesis from generated antisense RNA was performed using indirect labeling with CyScribe cDNA post labeling kit (Amersham Pharmacia Biotec, Buckinghamshire, England) according to the manufacturer’s instructions in the presence of fluorescent Cy3 and Cy5 dyes. After purification and denaturation, the labeled targets were hybridized to the microarrays at 55°C overnight. After hybridization, the arrays were washed under stringent conditions to remove unspecific target binding and were subsequently air dried. Fluorescence signals were analyzed using an Affymetrix 418 Array™ Scanner with the Imagen® software Version 3.0. The expression level was deduced from the ratio of signal intensity (Cy5/Cy3).
Data Preprocessing and Analysis
For each spot, median signal and background intensities for both channels were obtained. To account for spot differences, the background-corrected ratio of the two channels was calculated. Following the annotation of Yang et al. (92), we used the log M = log2 R/G and the mean log intensity A = log2√RG, and the MA plots as described by Dudoit et al. (16). Here, R and G denote the measured fluorescence intensities after background subtraction for the probes labeled with the Cy5 and Cy3 dyes, respectively.
To balance the fluorescence intensities for the two dyes as well as to allow the comparison of expression levels across experiments, the raw data were standardized. First, we used an intensity-dependent standardization as described by Yang et al. (92) to correct for inherent and random bias on each chip. In a second step, a global standardization was applied to center the log ratios at zero. As each gene was spotted twice on the chip, and four arrays were analyzed, mean log ratios M were calculated for each gene. A cutoff of threefold expression in the mean log ratio was chosen to identify candidate genes for differentially expression.
Direct Sequencing Analysis
Spots with differentially regulated genes were corroborated using sequencing analysis. To this end, plasmids with cDNA inserts from Research Genetics library were first cultivated inside E. coli with LB medium and then DNA extracted using the QIAprep® Spin Miniprep kit from Qiagen (Hilden, Germany) according to the manufacturer’s instructions. Sequencing was performed with a DNA sequencing kit (Applied Biosystems, Warrington, UK) as described by the manufacturer on an ABI PRISM™ 377 DNA sequencing device (Applied Biosystems).
Real-Time PCR Analysis
For selected genes, a quantitative expression analysis was employed. RNA was extracted from MSC and skin fibroblasts using a commercial kit (RNeasy Qiagen) according to the manufacturer’s instructions. From the same RNA previously used for preamplification and array hybridization, 1 μg RNA was taken for cDNA synthesis with the Omniscript™ RT-PCR kit from Qiagen as recommended by the manufacturer. β-Actin PCR was performed as control for the quality of cDNA on a thermal cycler PE 9600 (Applied Biosystems) as previously described (76), followed by visualization on an agarose gel. To quantify cDNA levels the QuantiTect™ SYBR® Green PCR kit (Qiagen) and 1 μl cDNA were used on an ABI PRISM 7700 Sequence Detector (Taqman™, Applied Biosystems) with the following amplification conditions: 45 cycles of three-step PCR; 94°C for 15 s, 56°C for 30 s, 72°C for 30 s for IGF2 and PPARGC1 primers and 94°C for 15 s, 58°C for 30 s, 72°C for 30 s for IGFBP5, LIM, and DKK3 primers after initial denaturation at 95°C for 15 min with QuantiTect™ SYBR® Green PCR kit (Qiagen) and 1μ l cDNA. Primer sequences are listed in Table 1. RNA levels were normalized using the β-actin housekeeping gene as described.
TABLE 1.
PRIMER SEQUENCES EMPLOYED IN REAL-TIME QUANTITATIVE PCR ANALYSIS OF FIVE SELECTED GENES THAT WERE FOUND TO BE REGULATED DIFFERENTLY IN MESENCHYMAL STEM CELLS COMPARED TO SKIN FIBROBLASTS
| Accession Number | Gene Name | Primer Sequence | Product Size (bp) |
|---|---|---|---|
| N74623 | Insulin-like growth factor 2 | forward: 5′-TTTCCGCAGCTGT-GACCTG-3′ | 134 |
| reverse: 5′-ATTGGAAGAACTTGCCCACG-3′ | |||
| N89673 | Peroxisome proliferative activated receptor γ | forward: 5′-TCAAATGAACAC GTGCACCC-3′ | 140 |
| reverse: 5′-AAAGCACCAGTTCGGTTACCA-3′ | |||
| T52830 | Human insulin-like growth factor binding protein 5 | forward: 5′-CAGTGCAAACCTTCCCGTG-3′ | 72 |
| reverse: 5′-TGGCAGCTTCATCCCGTACT-3′ | |||
| R92455 | LIM protein | forward: 5′-TCCAGCAGGGAAACGAACTC-3′ | 103 |
| reverse: 5′-GTTGAATTCT-TCTGGGTGCCA-3′ | |||
| AA425947 | Dickkopf homolog3 | forward: 5′-TCTGGACCTCATCACCTGGG-3′ | 105 |
| reverse: 5′-ACATACACCAGGCTGTGGCTG-3′ |
The accession number refers to the gen ID for GenBank research.
RESULTS
Human Bone Marrow Mesenchymal Cells Exhibit High Proliferation and Differentiation Abilities Compared to Skin Fibroblasts
To ensure that bone marrow-derived mesenchymal cells are true stem cells with capacity for self-renewal and differentiation, MSC were grown continuously up to 20–25 passages without loss of proliferation ability, viability, or differentiation capacity. Purity of MSC was confirmed by flow cytometric analysis: no hematopoietic cell, neither macrophages (CD14, CD45), blood progenitor cells (CD34, CD45), nor endothelial precursors (CD34, CD31) were detected (Fig. 1). Cloning efficiency of MSC as indicated by their ability to form CFU-F was checked in early and later cell passages and their potential to differentiate into osteocytes, chondrocytes, and adipocytes was tested in vitro. Skin fibroblasts were considered as mature counterparts and therefore also tested for their differentiation potential. These cells took considerably longer to grow confluent and could only be cultured for up to four passages. Therefore, we conclude that MSC but not skin fibroblasts resemble a stem cell phenotype with high proliferation capacity.
Figure 1.
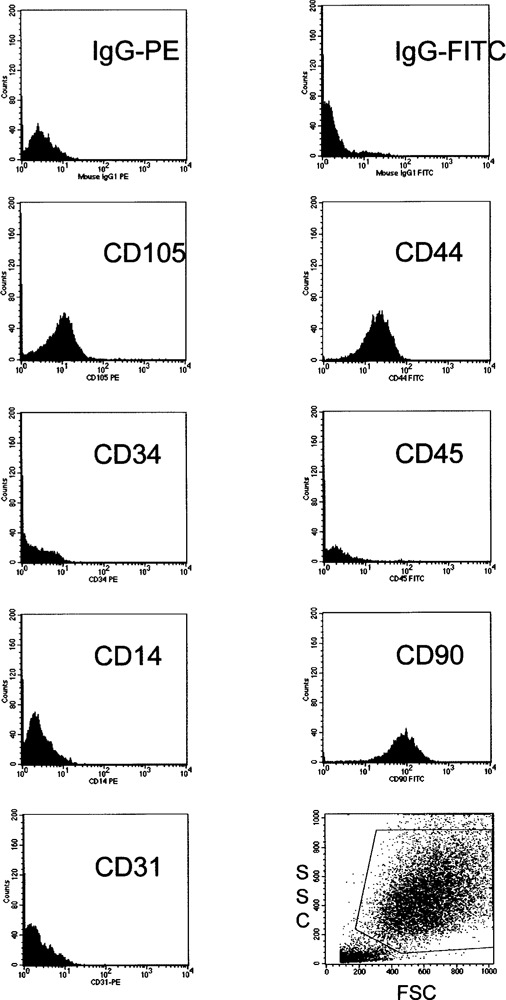
Homogeneous antigen profile of mesenchymal stem cells. Mesenchymal stem cells show homogeneous surface expression of CD44, CD90, and CD105. Staining for hematopoietic (CD45, CD14, CD34) and endothelial (CD34, CD31) cells was negative, thus proving purity and homogeneity of mesenchymal stem cell cultures.
MSC exhibit a high level of leucocyte-alkaline phosphatase (LAP) and bone sialoprotein on flow cytometric analysis as well as increased extracellular calcium deposit (Kossa staining) after induction of osteogenic differentiation (Fig. 2A, B). However, no or only very light staining with LAP, bone sialoprotein, or extracellular calcium deposits was detected in fibroblasts derived from human skin (Fig. 2C). Thus, MSC, but not skin fibroblasts, exhibited the ability for ex vivo osteogenic differentiation. This was also true for chondrogenic differentiation.
Figure 2.
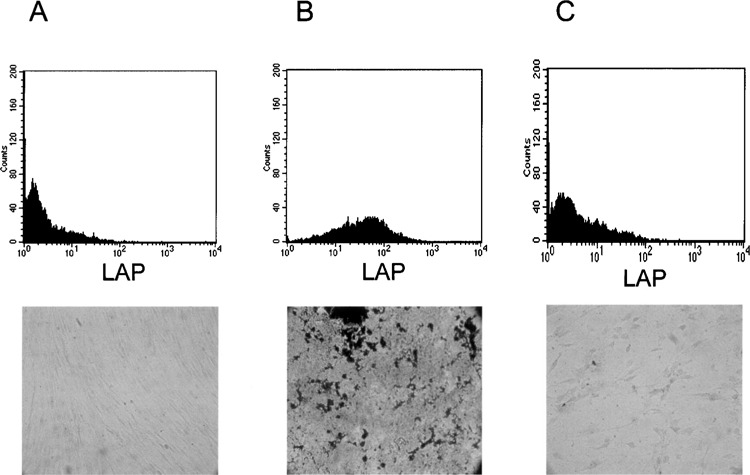
Mesenchymal stem cells but not skin fibroblasts harbor osteogenic differentiation abilities. Mesenchymal stem cells without induction of osteogenesis show no expression of alkaline phosphatase (LAP) by FACS analysis or calcium deposits in von Kossa staining (A). After induction of bone differentiation elevated LAP levels and a significant amount of calcium deposits can be detected in former mesenchymal stem cells (B), but not in skin fibroblasts (C).
Adipogenic differentiation capacity of MSC was demonstrated by increasing number of lipid vesicles, stained with Sudan black, but no fat vacuole was detected within the skin fibroblast culture. PCR analysis of the PPARγ2 gene, which is typically expressed in fat tissue, confirmed the adipogenic differentiation of MSC.
Therefore, we concluded that only MSC grown under standard conditions harbor stem cell plasticity, whereas skin fibroblast cannot be driven to differentiate into other mesenchymal lineages, indicating their mature state as skin stromal cells.
Mesenchymal Stem Cells Harbor Specific Gene Expression Signatures
Upon cDNA microarray analysis differential gene expression levels in MSC compared to mature fibroblasts was detected in about 80 from 9600 analyzed genes. Only genes with more than threefold difference in the expression level were considered for analysis.
Our data show that typical connective tissue genes encoding proteins like matrilin or extracellular matrix protein-1 (33,54) often had lower copy numbers in MSC versus skin fibroblast (Table 2), confirming the fibroblast progenitor status of MSC. In concordance with this finding, genes that occur in mature mesoderm-derived tissues like bone, fat, smooth muscle, or endothelium were often found to be downregulated in MSC [i.e., AE-binding protein-1 (74), the complement-regulatory gene decay-accelerating factor (DAF) (50), fatty acid binding protein 3, and PPARγ coactivator 1 (18,38,71)] (Table 2).
TABLE 2.
GENES THAT WERE FOUND TO BE UPREGULATED AND DOWNREGULATED IN MSC VERSUS SKIN FIBROBLASTS
| Expression Level | Accession Number | Gene Name | Function (References) |
|---|---|---|---|
| Upregulated genes | |||
| 10.0 | AA430540 | Collagen, type IV, alpha 2 | extracellular matrix protein |
| 8.8 | N74623 | Insulin-like growth factor-2 | regulation of normal cell growth (59) |
| 7.2 | AA136707 | Procollagene-lysine, 2-oxoglutarate 5-dioxygenase | collagen synthesis |
| 6.0 | AA459308 | Elastin | protein that provides the property of elastic recoil in dermis, lungs, and blood vessels (67) |
| 5.6 | N74882 | Distal-less homeobox 5 | organ development, limb initiation (21,95) |
| 5.4 | AA460975 | Scrapie responsive protein 1 | expressed by cells of neural origin in mouse embryo (15) |
| 4.6 | AA482119 | Inhibitor of DNA binding 3 | plays a role in keeping precursor cells immature (94) |
| 4.5 | T52830 | Human insulin-like growth factor binding protein 5 | growth and differentiation (26,53) |
| 4.4 | AA421819 | Cadherin 6, type 2, K-cadherin | cell adhesion molecule, expressed in normal kidney and renal cell carcinoma (65) |
| 4.0 | W49619 | Cadherin 2, type 1, N-cadherin | neural development (46,47) |
| 4.0 | AA487193 | Secreted frizzled-related protein 4 | transmembrane receptor; WNT network (19,34) |
| 4.0 | W72803 | EGF-TM7 latrophilin-related protein | transmembrane receptor, neutrophil migration (41) |
| 3.7 | R76614 | Netrin 4 | neuronal development (2,35,93) |
| 3.7 | AA074535 | Hematopoetic PBX-interacting protein | organization of cytoskeleton (1) |
| 3.4 | R92455 | LIM protein | angiogenesis and hematopoiesis (90,91) |
| 3.4 | AA402754 | Eyes absent (Drosophila) homolog 2 | dynamic expression during development (17) |
| 3.4 | H79023 | Disintegrin and metalloproteinase (meltrin α) ADAM 12 | disintegrin and metalloproteinase is expressed in human and rat brain, developing and regenerating heart, and skeletal muscle (7) |
| 3.3 | R55185 | EST, highly similar to IRX3 mouse II | IRX3 is involved in WNT signaling in developing neural tissue (9) and developing heart (12) |
| 3.3 | AA417279 | Protein tyrosine phosphatase, non-receptor type substrate 1 | expressed in rat kidney (55) |
| 3.1 | N79778 | Extracellular matrix protein 2 | extracellular matrix protein, female organ and adipocyte specific |
| 3.1 | R82176 | MAD (mothers against dpp = decapen-taplegic) homolog 7 | embryonic midgut development in drosophila; BMP-4 signaling pathway (58) |
| Downregulated genes | |||
| −30.3 | N32768 | Pregnancy specific beta-1-glycoprotein 3 | immunomodulating function (80) |
| −25.0 | W48852 | Cysteine knot superfamily 1 | growth factors (52) |
| −10.6 | R48303 | Dermatopontin | proteoglycan-binding cell adhesion protein, interacts with TGF-β (64) |
| −10.5 | AA496334 | Dynamin 1 | endocytosis (11) |
| −10.0 | AA430540 | Collagen, type IV, alpha 2 | extracellular matrix protein |
| −9.3 | AA148548 | Fatty acid binding protein 3 | fatty acid metabolism, trafficking, and signaling (71) |
| −8.7 | AA644088 | Cathepsin C | activates serin proteases like progranzymes (83,88) |
| −8.1 | N50845 | Contactin 3 | adhesion molecule, neural development (62,63) |
| −7.0 | H09748 | B-cell CLL/lymphoma 11B | zinc finger protein |
| −6.8 | T50121 | Kreisler maf-related leucine zipper homolog | transcription factor, activated HOXb-3 in segmental regulation (48) |
| −6.8 | W49781 | Leupaxin | phosphotyrosine protein most homologous to paxillin preferentially expressed in hematopoietic cells (45) |
| −6.3 | R66101 | Neuritin | neurogenesis (57) |
| −5.8 | W35153 | G protein-coupled receptor, family C, group 5, member B | |
| −5.7 | AA425947 | Dickkopf (Xenopus laevis) homolog 3 | candidate tumor suppressor gene (37,61) |
| −4.0 | N89673 | Peroxisome proliferative activated receptor γ coactivator 1 | fat metabolism, transcriptional coactivator (18,38) |
| −3.8 | AA490462 | AE-binding protein 1 | regulated in adipogenesis and vascular smooth muscle cell differentiation (74) |
| −3.6 | AA071473 | Matrilin 2 | development of cartilage and bones (23) |
| −3.6 | R09561 | Decay accelerating factor for complement | membrane-bound complement-regulatory protein (CD55, Cromer blood group system) (81) |
| −3.5 | N79484 | Extracellular matrix protein 1 | bone development and angiogenesis (33,54) |
| −3.4 | N23996 | SWAP-70 protein | Signaling of membrane ruffling (78) |
| −3.3 | N98485 | Forkhead box F2 | Potential transcriptional factor |
| −3.2 | H19315 | Contactin 1 | neural cell adhesion molecule |
Surprisingly, several genes of ectoderm origin, like contactin and G-protein-coupled receptor, which are considered as specific genes for neural tissues (10,75), were downregulated in MSC. In addition we found that cadherin 2 (46,47), scrapie responsive protein 1 (15), and netrin (2,35,93), which are all detected in developmental neural tissue, were expressed in high amounts in MSC. Nevertheless, neuritin (57) and contactin (62,63), which seem to be involved in neural development as well, were downregulated in MSC.
Interestingly, several other genes that are somehow involved in embryogenesis and organ development were found to be regulated differently in MSC, corroborating the progenitor cell character of MSC for tissues of all dermal layers. All these embryonic and developmental genes are summarized in Table 3.
TABLE 3.
MESENCHYMAL STEM CELLS DISPLAY A DIFFERENTIAL EXPRESSION OF GENES INVOLVED IN EMBRYOGENESIS AND TISSUE DEVELOPMENT
| Upregulated in MSC | Downregulated in MSC |
|---|---|
| Insulin-like growth factor II | Neuritin |
| Netrin | Contactin 3 |
| LIM protein | Kreisler (Kmrl1) |
| Secreted Frizzled-related protein 4 | Extracellular matrix protein 1 |
| Inhibitor of DNA binding 3 | |
| Distal-less homeo box 5 | |
| Scrapie responsive protein 1 | |
| Human insulin-like growth factor BP5 | |
| Cadherin 2, type 1 | |
| Eyes absent homolog 2 | |
| MAD homolog 7 |
We also detected about 30 ESTs (expressed sequenced tags) and some other genes like dickkopf-3 (Dkk-3/REIC), a tumor suppressor gene whose promoter is often methylated in human tumor cells (37,61), that was found to be downregulated in MSC (Table 2).
Direct Sequencing and Quantitative PCR Analysis Revealed Correct Gene Identity and Expression Quantification in MSC
Out of the 80 differently expressed genes 45 clones were selected randomly and their identity was confirmed by direct sequencing (Table 2). From the 45 sequenced genes one was a mixed clone and another one revealed to be another gene (i.e., it did not match the array spot). Therefore, the quality of our custom-made array chip proved to be valid and reliable regarding the identified genes.
In addition, real-time quantitative PCR analysis revealed a good correlation to the quantity of gene expression measured by our gene array system. We performed quantitative PCR analysis for five representative genes and demonstrated an equal expression level of the identified genes (Table 4). Interestingly, the level of up- or downregulation seemed to be even more pronounced employing real-time quantitative PCR analysis. Thus, our array data were confirmed and proved to be specific and sensitive without over-estimation of expression differences.
TABLE 4.
VERIFICATION OF GENE ARRAY RESULTS BY REAL-TIME QUANTITATIVE PCR ANALYSIS
| Gene (Accession No.) | Relative Gene Expression | |
|---|---|---|
| cDNA Array | Real-Time PCR | |
| Peroxisome proliferative activated receptor γ (N89673) | −4.0 | −6.6 |
| Dickkopf homolog 3 (AA425947) | −5.7 | −5.4 |
| LIM protein (R92455) | 3.4 | 12.2 |
| Insulin-like growth factor-2 (N74623) | 8.8 | 91.7 |
| Human insulin-like growth factor binding protein 5 (T52830) | 4.5 | 10.3 |
Gene expression measured with both techniques shows good correlation. Differences in expression levels seem to be more pronounced with real-time PCR analysis.
DISCUSSION
Although human bone marrow mesenchymal stem cells have a similar morphology and resemble the same immunophenotype such as human skin fibroblasts, distinct biological differences can be found with regard to growth pattern and differentiation capabilities. In order to address whether this is paralleled by different gene expression profiles we hybridized mature and immature fibroblastic cell types against each other. Therefore, in our approach genes and signaling pathways conferring stem cell abilities were likely to be revealed in MSC rather than typical fibroblastic lineage genes, like genes encoding for vimentin, different collagens, stromal cell-derived factor 1, or fibronectin. In concordance with this we found that those genes encoding for structural proteins typically active in mature mesodermal tissues were expressed in lower amounts in MSC (i.e., extracellular matrix protein 1, fatty acid binding protein 3, peroxisome proliferative activated receptor γ coactivator 1, AE-binding protein 1, or matrilin 2).
Interestingly, we discovered several differently regulated genes that play a role in neuroectodermal development: G-protein-coupled receptor and contactin, genes found in mature neural tissue (10,75), were downregulated in MSC. In contrast netrin, cadherin 2, and scrapie responsive protein 1, which can typically be found in developing neural tissue, were found to be upregulated in MSC. From these data we conclude a neuronal precursor status of MSC, which is confirmed by the finding of human adult MSC differentiating along neuronal pathway in vitro (32,36,42,70). In concordance with other groups, who found nestin and neurofilament H expressed in MSC (84), all these data confirm the ubiquitous germline, ectodermal, endodermal, and mesodermal gene expression profile of MSC (89).
Beyond that, we were able to demonstrate that MSC regulate a variety of genes that are abundantly expressed in the early embryo or during organ development, although MSC are considered to be “adult” stem cells. Most of these developmental genes were found to be upregulated, such as distal-less homeo box 5 gene, which plays a role in embryonic organ development (95) and especially limb initiation (21), but a few of them were downregulated as well, as depicted in Table 3. Pronounced expression of developmental genes in MSC was corroborated by another group comparing gene expression of MSC versus CD34 (79) and indicates inherent plasticity and potential remodeling facilities of MSC.
Although results of gene array printings vary considerably because of different stem cell sources, stem cell purity (87), different array chips probed, or statistical analysis employed in data interpretation (20,22), identification of common “stem cell genes” in two or more studies is statistically significant and warrants further investigation. In our approach we identified five genes that were already described as “stem cell” genes before by comparing the gene expression profile of hematopoietic, neural, and embryonic cells by microarray analysis (30,72).
Among the proteins encoded by these enhanced genes were four and a half LIM protein, which plays a pivotal role in yolk sac erythropoiesis and in the proper development of all hematopoietic lineages in the adult individual (91) as well as angiogenesis (90) and interacts with human insulin-like growth factor binding protein 5 (IGFBP-5) (3) (Table 3). IGFBP-5 is the most conserved IGFBP across species among the family of IGF binding proteins (IGFBP-1 to -6), and was identified as an essential regulator of physiological processes in bone, kidney, and mammary gland (6,14,49,77,86). In addition, IGFBPs appear to play a decisive role in control of proliferation of specific tumor cell lines (44,82) as well as limb development (53) and growth of smooth muscle cells (39).
The distal-less homeo box 5 gene was also described as a “stem cell” gene before (30) and was found to be enriched in our MSC. Its upregulation seems to be a unique stem cell feature, because it is found in four different stem cell entities.
In addition we identified two MSC “stemness” genes of the Wnt signaling network, which is involved in embryo development and cancer formation [i.e., the gene for secreted frizzled-related protein 4, a protein abundantly expressed in the early embryo (30,34) and dickkopf-3 (dkk-3), related to Dickkopf protein 1 (dkk1) (34)]. dickkopf-3 also resembles a candidate tumor suppressor gene responsible for aging processes (37,61,85). In summary, these regulators of this Wnt/β-catenin pathway seem to play a prominent role in MSC, because they have been described as an autocrine signaling mechanism operating in MSC by different investigators (19) also.
Although the precise signaling pathways that determine the differentiated fate of MSC is not fully understood yet, our data confirm the idea that, unlike skin fibroblasts, adult bone marrow mesenchymal stem cells in humans may have true stem cell characteristics. Our finding is in concordance with data from other groups, who approached the gene expression profiling in mesenchymal stem cells upon in vitro differentiation into osteogenic, chondrogenic, and adipogenic tissue (4,13,28,29,40,60), but only one to three overlapping genes are confirmed in each gene set employed. The issue of reproducibility of gene expression profiles under different conditions (culture passages, media and serum conditions, array slides) has been discussed extensively (20,22,40,87).
In conclusion, no single study can confidently identify bona fide stem cell genes; cross-validation of gene array results generated independently by different investigators remains to be crucial. With our approach we were able to confirm gene expression profiles that were described for stem cells and developmental processes before, and future studies may reveal more details in signaling pathways responsible for the respective differentiation processes.
ACKNOWLEDGMENTS
The authors would like to thank the German Bundesministerium für Bildung und Forschung (BMBF; grant 01GN0125) and Medac Schering Oncology GmbH for financial support. We also thank Prof. Eilers from the Institute of Molecular Biology and Tumorforschung (IMT) Marburg, Germany, for the supportive co-work of his department and the Kempkes Stiftung Marburg for initial financial support to start the work about MSC in our department.
REFERENCES
- 1. Abramovich C.; Chavez E. A.; Lansdorp P. M.; Humphries R. K. Functional characterization of multiple domains involved in the subcellular localization of the hematopoietic Pbx interacting protein (HPIP). Oncogene 21:6766–6771; 2002. [DOI] [PubMed] [Google Scholar]
- 2. Aisemberg G. O.; Kuhn J.; Macagno E. R. Netrin signal is produced in leech embryos by segmentally iterated sets of central neurons and longitudinal muscle cells. Dev. Genes Evol. 211:589–596; 2001. [DOI] [PubMed] [Google Scholar]
- 3. Amaar Y. G.; Thompson G. R.; Linkhart T. A.; Chen S. T.; Baylink D. J.; Mohan S. Insulin-like growth factor-binding protein 5 (IGFBP-5) interacts with a four and a half LIM protein 2 (FHL2). J. Biol. Chem. 277:12053–12060; 2002. [DOI] [PubMed] [Google Scholar]
- 4. Barry F.; Boynton R. E.; Liu B.; Murphy J. M. Chondrogenic differentiation of mesenchymal stem cells from bone marrow: Differentiation-dependent gene expression of matrix components. Exp. Cell Res. 268:189–200; 2001. [DOI] [PubMed] [Google Scholar]
- 5. Baugh L. R.; Hill A. A.; Brown E. L.; Hunter C. P. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 29:E29; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berfield A. K.; Andress D. L.; Abrass C. K. IGFBP-5(201-218) stimulates Cdc42GAP aggregation and filopodia formation in migrating mesangial cells. Kidney Int. 57:1991–2003; 2000. [DOI] [PubMed] [Google Scholar]
- 7. Bernstein H. G.; Keilhoff G.; Bukowska A.; Ziegeler A.; Funke S.; Dobrowolny H.; Kanakis D.; Bogerts B.; Lendeckel U. ADAM (a disintegrin and metalloprotease) 12 is expressed in rat and human brain and localized to oligodendrocytes. J. Neurosci. Res. 75:353–360; 2004. [DOI] [PubMed] [Google Scholar]
- 8. Berwanger B.; Hartmann O.; Bergmann E.; Bernard S.; Nielsen D.; Krause M.; Kartal A.; Flynn D.; Wiedemeyer R.; Schwab M.; Schafer H.; Christiansen H.; Eilers M. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell 2:377–386; 2002. [DOI] [PubMed] [Google Scholar]
- 9. Braun M. M.; Etheridge A.; Bernard A.; Robertson C. P.; Roelink H. Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development 130:5579–5587; 2003. [DOI] [PubMed] [Google Scholar]
- 10. Brauner-Osborne H.; Krogsgaard-Larsen P. Sequence and expression pattern of a novel human orphan G-protein-coupled receptor, GPRC5B, a family C receptor with a short amino-terminal domain. Genomics 65:121–128; 2000. [DOI] [PubMed] [Google Scholar]
- 11. Choi J. H.; Park J. B.; Bae S. S.; Yun S.; Kim H. S.; Hong W. P.; Kim I. S.; Kim J. H.; Han M. Y.; Ryu S. H.; Patterson R. L.; Snyder S. H.; Suh P. G. Phospholipase C-gamma1 is a guanine nucleotide exchange factor for dynamin-1 and enhances dynamin-1-dependent epidermal growth factor receptor endocytosis. J. Cell Sci. 117:3785–3795; 2004. [DOI] [PubMed] [Google Scholar]
- 12. Christoffels V. M.; Keijser A. G.; Houweling A. C.; Clout D. E.; Moorman A. F. Patterning the embryonic heart: Identification of five mouse Iroquois homeobox genes in the developing heart. Dev. Biol. 224:263–274; 2000. [DOI] [PubMed] [Google Scholar]
- 13. de Jong D. S.; Vaes B. L.; Dechering K. J.; Feijen A.; Hendriks J. M.; Wehrens R.; Mummery C. L.; van Zoelen E. J.; Olijve W.; Steegenga W. T. Identification of novel regulators associated with early-phase osteoblast differentiation. J. Bone Miner. Res. 19:947–958; 2004. [DOI] [PubMed] [Google Scholar]
- 14. Devlin R. D.; Du Z.; Buccilli V.; Jorgetti V.; Canalis E. Transgenic mice overexpressing insulin-like growth factor binding protein-5 display transiently decreased osteoblastic function and osteopenia. Endocrinology 143:3955–3962; 2002. [DOI] [PubMed] [Google Scholar]
- 15. Dron M.; Tartare X.; Guillo F.; Haik S.; Barbin G.; Maury C.; Tovey M.; Dandoy-Dron F. Mouse scrapie responsive gene 1 (Scrg1): Genomic organization, physical linkage to sap30, genetic mapping on chromosome 8, and expression in neuronal primary cell cultures. Genomics 70:140–149; 2000. [DOI] [PubMed] [Google Scholar]
- 16. Dudoit S.; Fridlyand J. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol. 3:RESEARCH0036; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duncan M. K.; Kos L.; Jenkins N. A.; Gilbert D. J.; Copeland N. G.; Tomarev S. I. Eyes absent: A gene family found in several metazoan phyla. Mamm. Genome 8:479–485; 1997. [DOI] [PubMed] [Google Scholar]
- 18. Esterbauer H.; Oberkofler H.; Linnemayr V.; Iglseder B.; Hedegger M.; Wolfsgruber P.; Paulweber B.; Fastner G.; Krempler F.; Patsch W. Peroxisome proliferator-activated receptor-gamma coactivator-1 gene locus: Associations with obesity indices in middle-aged women. Diabetes 51:1281–1286; 2002. [DOI] [PubMed] [Google Scholar]
- 19. Etheridge S. L.; Spencer G. J.; Heath D. J.; Genever P. G. Expression profiling and functional analysis of wnt signaling mechanisms in mesenchymal stem cells. Stem Cells 22:849–860; 2004. [DOI] [PubMed] [Google Scholar]
- 20. Evsikov A. V.; Solter D. Comment on “‘Stemness’: Transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature.” Science 302:393; author reply 393; 2003. [DOI] [PubMed] [Google Scholar]
- 21. Ferrari D.; Harrington A.; Dealy C. N.; Kosher R. A. Dlx-5 in limb initiation in the chick embryo. Dev. Dyn. 216:10–15; 1999. [DOI] [PubMed] [Google Scholar]
- 22. Fortunel N. O.; Otu H. H.; Ng H. H.; Chen J.; Mu X.; Chevassut T.; Li X.; Joseph M.; Bailey C.; Hatzfeld J. A.; Hatzfeld A.; Usta F.; Vega V. B.; Long P. M.; Libermann T. A.; Lim B. Comment on “‘Stemness’: Transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature.” Science 302:393; author reply 393; 2003. [DOI] [PubMed] [Google Scholar]
- 23. Frank S.; Schulthess T.; Landwehr R.; Lustig A.; Mini T.; Jeno P.; Engel J.; Kammerer R. A. Characterization of the matrilin coiled-coil domains reveals seven novel isoforms. J. Biol. Chem. 277:19071–19079; 2002. [DOI] [PubMed] [Google Scholar]
- 24. Friedenstein A. J.; Gorskaja J. F.; Kulagina N. N. Fibroblast precursors in normal and irradiated mouse hematopoietic organs. Exp. Hematol. 4:267–274; 1976. [PubMed] [Google Scholar]
- 25. Friedenstein A. J.; Petrakova K. V.; Kurolesova A. I.; Frolova G. P. Heterotopic transplants of bone marrow: Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation 6:230–247; 1968. [PubMed] [Google Scholar]
- 26. Giudice L. C.; Conover C. A.; Bale L.; Faessen G. H.; Ilg K.; Sun I.; Imani B.; Suen L. F.; Irwin J. C.; Christiansen M.; Overgaard M. T.; Oxvig C. Identification and regulation of the IGFBP-4 protease and its physiological inhibitor in human trophoblasts and endometrial stroma: Evidence for paracrine regulation of IGF-II bioavailability in the placental bed during human implantation. J. Clin. Endocrinol. Metab. 87:2359–2366; 2002. [DOI] [PubMed] [Google Scholar]
- 27. Gorski J. P. Acidic phosphoproteins from bone matrix: A structural rationalization of their role in biomineralization. Calcif. Tissue Int. 50:391–396; 1992. [DOI] [PubMed] [Google Scholar]
- 28. Hung S. C.; Chang C. F.; Ma H. L.; Chen T. H.; Low-Tone Ho L. Gene expression profiles of early adipogenesis in human mesenchymal stem cells. Gene 340:141–150; 2004. [DOI] [PubMed] [Google Scholar]
- 29. Imabayashi H.; Mori T.; Gojo S.; Kiyono T.; Sugiyama T.; Irie R.; Isogai T.; Hata J.; Toyama Y.; Umezawa A. Redifferentiation of dedifferentiated chondrocytes and chondrogenesis of human bone marrow stromal cells via chondrosphere formation with expression profiling by large-scale cDNA analysis. Exp. Cell Res. 288:35–50; 2003. [DOI] [PubMed] [Google Scholar]
- 30. Ivanova N. B.; Dimos J. T.; Schaniel C.; Hackney J. A.; Moore K. A.; Lemischka I. R. A stem cell molecular signature. Science 298:601–604; 2002. [DOI] [PubMed] [Google Scholar]
- 31. Jiang Y.; Jahagirdar B. N.; Reinhardt R. L.; Schwartz R. E.; Keene C. D.; Ortiz-Gonzalez X. R.; Reyes M.; Lenvik T.; Lund T.; Blackstad M.; Du J.; Aldrich S.; Lisberg A.; Low W. C.; Largaespada D. A.; Verfaillie C. M. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 20:1–9; 2002. [DOI] [PubMed] [Google Scholar]
- 32. Joannides A.; Gaughwin P.; Scott M.; Watt S.; Compston A.; Chandran S. Postnatal astrocytes promote neural induction from adult human bone marrow-derived stem cells. J. Hematother. Stem Cell Res. 12:681–688; 2003. [DOI] [PubMed] [Google Scholar]
- 33. Johnson M. R.; Wilkin D. J.; Vos H. L.; Ortiz de Luna R. I.; Dehejia A. M.; Polymeropoulos M. H.; Francomano C. A. Characterization of the human extracellular matrix protein 1 gene on chromosome 1q21. Matrix Biol. 16:289–292; 1997. [DOI] [PubMed] [Google Scholar]
- 34. Jones S. E.; Jomary C. Secreted Frizzled-related proteins: Searching for relationships and patterns. Bioessays 24:811–820; 2002. [DOI] [PubMed] [Google Scholar]
- 35. Kennedy T. E. Cellular mechanisms of netrin function: Long-range and short-range actions. Biochem. Cell. Biol. 78:569–575; 2000. [PubMed] [Google Scholar]
- 36. Kim B. J.; Seo J. H.; Bubien J. K.; Oh Y. S. Differentiation of adult bone marrow stem cells into neuroprogenitor cells in vitro. Neuroreport 13:1185–1188; 2002. [DOI] [PubMed] [Google Scholar]
- 37. Kobayashi K.; Ouchida M.; Tsuji T.; Hanafusa H.; Miyazaki M.; Namba M.; Shimizu N.; Shimizu K. Reduced expression of the REIC/Dkk-3 gene by promoter-hypermethylation in human tumor cells. Gene 282:151–158; 2002. [DOI] [PubMed] [Google Scholar]
- 38. Krempler F.; Breban D.; Oberkofler H.; Esterbauer H.; Hell E.; Paulweber B.; Patsch W. Leptin, peroxisome proliferator-activated receptor-gamma, and CCAAT/ enhancer binding protein-alpha mRNA expression in adipose tissue of humans and their relation to cardiovascular risk factors. Arterioscler. Thromb. Vasc. Biol. 20:443–449; 2000. [DOI] [PubMed] [Google Scholar]
- 39. Kuemmerle J. F.; Zhou H. Insulin-like growth factor-binding protein-5 (IGFBP-5) stimulates growth and IGF-I secretion in human intestinal smooth muscle by Rasdependent activation of p38 MAP kinase and Erk1/2 pathways. J. Biol. Chem. 277:20563–20571; 2002. [DOI] [PubMed] [Google Scholar]
- 40. Lee R. H.; Kim B.; Choi I.; Kim H.; Choi H. S.; Suh K.; Bae Y. C.; Jung J. S. Characterization and expression analysis of mesenchymal stem cells from human bone marrow and adipose tissue. Cell Physiol. Biochem. 14:311–324; 2004. [DOI] [PubMed] [Google Scholar]
- 41. Leemans J. C.; te Velde A. A.; Florquin S.; Bennink R. J.; de Bruin K.; van Lier R. A.; van der Poll T.; Hamann J. The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. J. Immunol. 172:1125–1131; 2004. [DOI] [PubMed] [Google Scholar]
- 42. Levy Y. S.; Merims D.; Panet H.; Barhum Y.; Melamed E.; Offen D. Induction of neuron-specific enolase promoter and neuronal markers in differentiated mouse bone marrow stromal cells. J. Mol. Neurosci. 21:121–132; 2003. [DOI] [PubMed] [Google Scholar]
- 43. Liechty K. W.; MacKenzie T. C.; Shaaban A. F.; Radu A.; Moseley A. M.; Deans R.; Marshak D. R.; Flake A. W. Human mesenchymal stem cells engraft and demonstrate site-specific differentiation after in utero transplantation in sheep. Nat. Med. 6:1282–1286; 2000. [DOI] [PubMed] [Google Scholar]
- 44. Lin S. C.; Wang C. P.; Chen Y. M.; Lu S. Y.; Fann M. J.; Liu C. J.; Kao S. Y.; Chang K. W. Regulation of IGFBP-5 expression during tumourigenesis and differentiation of oral keratinocytes. J. Pathol. 198:317–125; 2002. [DOI] [PubMed] [Google Scholar]
- 45. Lipsky B. P.; Beals C. R.; Staunton D. E. Leupaxin is a novel LIM domain protein that forms a complex with PYK2. J. Biol. Chem. 273:11709–11713; 1998. [DOI] [PubMed] [Google Scholar]
- 46. Liu Q.; Babb S. G.; Novince Z. M.; Doedens A. L.; Marrs J.; Raymond P. A. Differential expression of cadherin-2 and cadherin-4 in the developing and adult zebrafish visual system. Vis. Neurosci. 18:923–933; 2001. [PubMed] [Google Scholar]
- 47. Ludwig D.; Lorenz J.; Dejana E.; Bohlen P.; Hicklin D. J.; Witte L.; Pytowski B. cDNA cloning, chromosomal mapping, and expression analysis of human VE-Cadherin-2. Mamm. Genome 11:1030–1033; 2000. [DOI] [PubMed] [Google Scholar]
- 48. Manzanares M.; Cordes S.; Kwan C. T.; Sham M. H.; Barsh G. S.; Krumlauf R. Segmental regulation of Hoxb-3 by kreisler. Nature 387:191–195; 1997. [DOI] [PubMed] [Google Scholar]
- 49. Marshman E.; Green K. A.; Flint D. J.; White A.; Streuli C. H.; Westwood M. Insulin-like growth factor binding protein 5 and apoptosis in mammary epithelial cells. J. Cell Sci. 116:675–682; 2003. [DOI] [PubMed] [Google Scholar]
- 50. Mason J. C.; Lidington E. A.; Ahmad S. R.; Haskard D. O. bFGF and VEGF synergistically enhance endothelial cytoprotection via decay-accelerating factor induction. Am. J. Physiol. Cell Physiol. 282:C578–587; 2002. [DOI] [PubMed] [Google Scholar]
- 51. Masuhara K.; Suzuki S.; Yoshikawa H.; Tsuda T.; Takaoka K.; Ono K.; Morris D. C.; Hsu H. H.; Anderson H. C. Development of a monoclonal antibody specific for human bone alkaline phosphatase. Bone Miner. 17:182–186; 1992. [DOI] [PubMed] [Google Scholar]
- 52. McDonald N. Q.; Hendrickson W. A. A structural superfamily of growth factors containing a cystine knot motif. Cell 73:421–424; 1993. [DOI] [PubMed] [Google Scholar]
- 53. McQueeney K.; Dealy C. N. Roles of insulin-like growth factor-I (IGF-I) and IGF-I binding protein-2 (IGFBP2) and -5 (IGFBP5) in developing chick limbs. Growth Horm. IGF Res. 11:346–363; 2001. [DOI] [PubMed] [Google Scholar]
- 54. Mongiat M.; Fu J.; Oldershaw R.; Greenhalgh R.; Gown A. M.; Iozzo R. V. Perlecan protein core interacts with extracellular matrix protein 1 (ECM1), a glycoprotein involved in bone formation and angiogenesis. J. Biol. Chem. 278:17491–17499; 2003. [DOI] [PubMed] [Google Scholar]
- 55. Moriyama T.; Kawanishi S.; Inoue T.; Imai E.; Kaneko T.; Xia C.; Takenaka M.; Noguchi T.; Kamada T.; Ueda N. cDNA cloning of a cytosolic protein tyrosine phosphatase (RKPTP) from rat kidney. FEBS Lett. 353:305–308; 1994. [DOI] [PubMed] [Google Scholar]
- 56. Morris D. C.; Masuhara K.; Takaoka K.; Ono K.; Anderson H. C. Immunolocalization of alkaline phosphatase in osteoblasts and matrix vesicles of human fetal bone. Bone Miner. 19:287–298; 1992. [DOI] [PubMed] [Google Scholar]
- 57. Naeve G. S.; Ramakrishnan M.; Kramer R.; Hevroni D.; Citri Y.; Theill L. E. Neuritin: A gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc. Natl. Acad. Sci. USA 94:2648–2653; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Newfeld S. J.; Chartoff E. H.; Graff J. M.; Melton D. A.; Gelbart W. M. Mothers against dpp encodes a conserved cytoplasmic protein required in DPP/TGF-beta responsive cells. Development 122:2099–2108; 1996. [DOI] [PubMed] [Google Scholar]
- 59. Nielsen F. C. The molecular and cellular biology of insulin-like growth factor II. Prog. Growth Factor Res. 4:257–290; 1992. [DOI] [PubMed] [Google Scholar]
- 60. Nishioka K.; Dennis J. E.; Gao J.; Goldberg V. M.; Caplan A. I. Sustained Wnt protein expression in chondral constructs from mesenchymal stem cells. J. Cell. Physiol. 203:6–14; 2004. [DOI] [PubMed] [Google Scholar]
- 61. Nozaki I.; Tsuji T.; Iijima O.; Ohmura Y.; Andou A.; Miyazaki M.; Shimizu N.; Namba M. Reduced expression of REIC/Dkk-3 gene in non-small cell lung cancer. Int. J. Oncol. 19:117–121; 2001. [DOI] [PubMed] [Google Scholar]
- 62. Ogawa J.; Kaneko H.; Masuda T.; Nagata S.; Hosoya H.; Watanabe K. Novel neural adhesion molecules in the Contactin/F3 subgroup of the immunoglobulin superfamily: Isolation and characterization of cDNAs from rat brain. Neurosci. Lett. 218:173–176; 1996. [DOI] [PubMed] [Google Scholar]
- 63. Ogawa J.; Lee S.; Itoh K.; Nagata S.; Machida T.; Takeda Y.; Watanabe K. Neural recognition molecule NB-2 of the contactin/F3 subgroup in rat: Specificity in neurite outgrowth-promoting activity and restricted expression in the brain regions. J. Neurosci. Res. 65:100–110; 2001. [DOI] [PubMed] [Google Scholar]
- 64. Okamoto O.; Fujiwara S.; Abe M.; Sato Y. Dermatopontin interacts with transforming growth factor beta and enhances its biological activity. Biochem. J. 337:537–541; 1999. [PMC free article] [PubMed] [Google Scholar]
- 65. Paul R.; Necknig U.; Busch R.; Ewing C. M.; Hartung R.; Isaacs W. B. Cadherin-6: A new prognostic marker for renal cell carcinoma. J. Urol. 171:97–101; 2004. [DOI] [PubMed] [Google Scholar]
- 66. Phillips J.; Eberwine J. H. Antisense RNA amplification: A linear amplification method for analyzing the mRNA population from single living cells. Methods 10:283–288; 1996. [DOI] [PubMed] [Google Scholar]
- 67. Piontkivska H.; Zhang Y.; Green E. D.; Elnitski L. Multi-species sequence comparison reveals dynamic evolution of the elastin gene that has involved purifying selection and lineage-specific insertions/deletions. BMC Genomics 5:31; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pittenger M. F.; Mackay A. M.; Beck S. C.; Jaiswal R. K.; Douglas R.; Mosca J. D.; Moorman M. A.; Simonetti D. W.; Craig S.; Marshak D. R. Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147; 1999. [DOI] [PubMed] [Google Scholar]
- 69. Prockop D. J. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 276:71–74; 1997. [DOI] [PubMed] [Google Scholar]
- 70. Qian L.; Saltzman W. M. Improving the expansion and neuronal differentiation of mesenchymal stem cells through culture surface modification. Biomaterials 25:1331–1337; 2004. [DOI] [PubMed] [Google Scholar]
- 71. Qian Q.; Kuo L.; Yu Y. T.; Rottman J. N. A concise promoter region of the heart fatty acid-binding protein gene dictates tissue-appropriate expression. Circ. Res. 84:276–289; 1999. [DOI] [PubMed] [Google Scholar]
- 72. Ramalho-Santos M.; Yoon S.; Matsuzaki Y.; Mulligan R. C.; Melton D. A. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science 298:597–600; 2002. [DOI] [PubMed] [Google Scholar]
- 73. Reyes M.; Lund T.; Lenvik T.; Aguiar D.; Koodie L.; Verfaillie C. M. Purification and ex vivo expansion of postnatal human marrow mesodermal progenitor cells. Blood 98:2615–2625; 2001. [DOI] [PubMed] [Google Scholar]
- 74. Ro H. S.; Kim S. W.; Wu D.; Webber C.; Nicholson T. E. Gene structure and expression of the mouse adipocyte enhancer-binding protein. Gene 280:123–133; 2001. [DOI] [PubMed] [Google Scholar]
- 75. Robbins M. J.; Charles K. J.; Harrison D. C.; Pangalos M. N. Localisation of the GPRC5B receptor in the rat brain and spinal cord. Brain Res. Mol. Brain Res. 106:136–144; 2002. [DOI] [PubMed] [Google Scholar]
- 76. Schmidt M.; Nagel S.; Proba J.; Thiede C.; Ritter M.; Waring J. E.; Rosenbauer F.; Huhn D.; Wittig B.; Horak I.; Neubauer A. Lack of interferon-consensus-sequence-binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 91:22–29; 1998. [PubMed] [Google Scholar]
- 77. Schneider M. R.; Wolf E.; Hoeflich A.; Lahm H. IGF-binding protein-5: Flexible player in the IGF system and effector on its own. J. Endocrinol. 172:423–440; 2002. [DOI] [PubMed] [Google Scholar]
- 78. Shinohara M.; Terada Y.; Iwamatsu A.; Shinohara A.; Mochizuki N.; Higuchi M.; Gotoh Y.; Ihara S.; Nagata S.; Itoh H.; Fukui Y.; Jessberger R. SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature 416:759–763; 2002. [DOI] [PubMed] [Google Scholar]
- 79. Silva W. A. Jr.; Covas D. T.; Panepucci R. A.; Proto-Siqueira R.; Siufi J. L.; Zanette D. L.; Santos A. R.; Zago M. A. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells 21:661–669; 2003. [DOI] [PubMed] [Google Scholar]
- 80. Snyder S. K.; Wessner D. H.; Wessells J. L.; Waterhouse R. M.; Wahl L. M.; Zimmermann W.; Dveksler G. S. Pregnancy-specific glycoproteins function as immunomodulators by inducing secretion of IL-10, IL-6 and TGF-beta1 by human monocytes. Am. J. Reprod. Immunol. 45:205–216; 2001. [DOI] [PubMed] [Google Scholar]
- 81. Storry J. R.; Reid M. E. The Cromer blood group system: A review. Immunohematology 18:95–103; 2002. [PubMed] [Google Scholar]
- 82. Tanno B.; Negroni A.; Vitali R.; Pirozzoli M. C.; Cesi V.; Mancini C.; Calabretta B.; Raschella G. Expression of insulin-like growth factor-binding protein 5 in neuroblastoma cells is regulated at the transcriptional level by c-Myb and B-Myb via direct and indirect mechanisms. J. Biol. Chem. 277:23172–23180; 2002. [DOI] [PubMed] [Google Scholar]
- 83. Tran T. V.; Ellis K. A.; Kam C. M.; Hudig D.; Powers J. C. Dipeptidyl peptidase I: Importance of progranzyme activation sequences, other dipeptide sequences, and the N-terminal amino group of synthetic substrates for enzyme activity. Arch. Biochem. Biophys. 403:160–170; 2002. [DOI] [PubMed] [Google Scholar]
- 84. Tremain N.; Korkko J.; Ibberson D.; Kopen G. C.; DiGirolamo C.; Phinney D. G. MicroSAGE analysis of 2,353 expressed genes in a single cell-derived colony of undifferentiated human mesenchymal stem cells reveals mRNAs of multiple cell lineages. Stem Cells 19:408–418; 2001. [DOI] [PubMed] [Google Scholar]
- 85. Tsuji T.; Miyazaki M.; Sakaguchi M.; Inoue Y.; Namba M. A REIC gene shows down-regulation in human immortalized cells and human tumor-derived cell lines. Biochem. Biophys. Res. Commun. 268:20–24; 2000. [DOI] [PubMed] [Google Scholar]
- 86. Ulinski T.; Mohan S.; Kiepe D.; Blum W. F.; Wingen A. M.; Mehls O.; Tonshoff B. Serum insulin-like growth factor binding protein (IGFBP)-4 and IGFBP-5 in children with chronic renal failure: Relationship to growth and glomerular filtration rate. The European Study Group for Nutritional Treatment of Chronic Renal Failure in Childhood. German Study Group for Growth Hormone Treatment in Chronic Renal Failure. Pediatr. Nephrol. 14:589–597; 2000. [DOI] [PubMed] [Google Scholar]
- 87. Vogel G. Stem cells. ‘Stemness’ genes still elusive. Science 302:371; 2003. [DOI] [PubMed] [Google Scholar]
- 88. Wolters P. J.; Pham C. T.; Muilenburg D. J.; Ley T. J.; Caughey G. H. Dipeptidyl peptidase I is essential for activation of mast cell chymases, but not tryptases, in mice. J. Biol. Chem. 276:18551–18556; 2001. [DOI] [PubMed] [Google Scholar]
- 89. Woodbury D.; Reynolds K.; Black I. B. Adult bone marrow stromal stem cells express germline, ectodermal, endodermal, and mesodermal genes prior to neurogenesis. J. Neurosci. Res. 69:908–917; 2002. [DOI] [PubMed] [Google Scholar]
- 90. Yamada Y.; Pannell R.; Forster A.; Rabbitts T. H. The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice. Proc. Natl. Acad. Sci. USA 97:320–324; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yamada Y.; Warren A. J.; Dobson C.; Forster A.; Pannell R.; Rabbitts T. H. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. USA 95:3890–3895; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yang Y. H.; Dudoit S.; Luu P.; Lin D. M.; Peng V.; Ngai J.; Speed T. P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yin Y.; Sanes J. R.; Miner J. H. Identification and expression of mouse netrin-4. Mech. Dev. 96:115–119; 2000. [DOI] [PubMed] [Google Scholar]
- 94. Yokota Y. Id and development. Oncogene 20:8290–8298; 2001. [DOI] [PubMed] [Google Scholar]
- 95. Zerucha T.; Ekker M. Distal-less-related homeobox genes of vertebrates: Evolution, function, and regulation. Biochem. Cell. Biol. 78:593–601; 2000. [PubMed] [Google Scholar]


