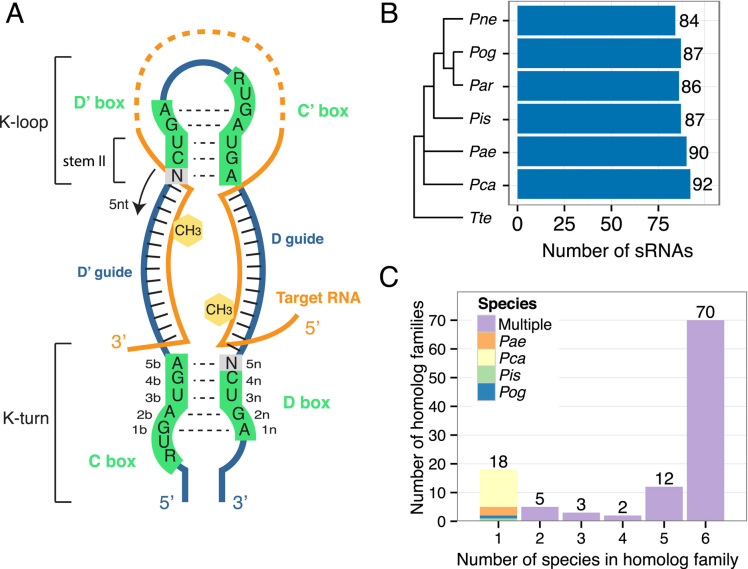
Figure 1.
Organization of Pyrobaculum C/D box RNAs into 110 homologous families. (A) The typical structure of an archaeal C/D box sRNA is depicted. The structure contains two K-motifs, the K-turn formed by the interaction between the C and D box sequences and the K-loop formed by the interaction between the D′ and C′ motifs (black dashed lines). The two guide regions located respectively between the D′ and C boxes and between the D and C′ boxes (green), base pair with the target RNA (orange) and methylation (yellow hexagon) occurs in the target nucleotide that base pairs with the guide 5 nts upstream from the start of the D′ or D box sequence. This is the ‘N+five’ rule. (B) The number of identified sRNAs in each of the six species of Pyrobaculum is indicated. Species are ordered based on a phylogenetic tree determined by 16S rRNA alignment (19). Thermoproteus tenax (Tte) is included as an outgroup. (C) C/D box sRNAs were organized into 110 homologous families based on sequence similarity of the guides and predicted targets in rRNA and tRNAs. C/D box sRNA numbers indicate to which family each belongs. Thus, Pae sR01, Par sR01, etc. belong to the sR01 family. C/D box sRNAs were first grouped into families using the original annotation numbering in Pae (1–65) (25). All other C/D box sRNAs were grouped into families starting at number 100. The majority of sRNAs fall into families with representatives in all six species.