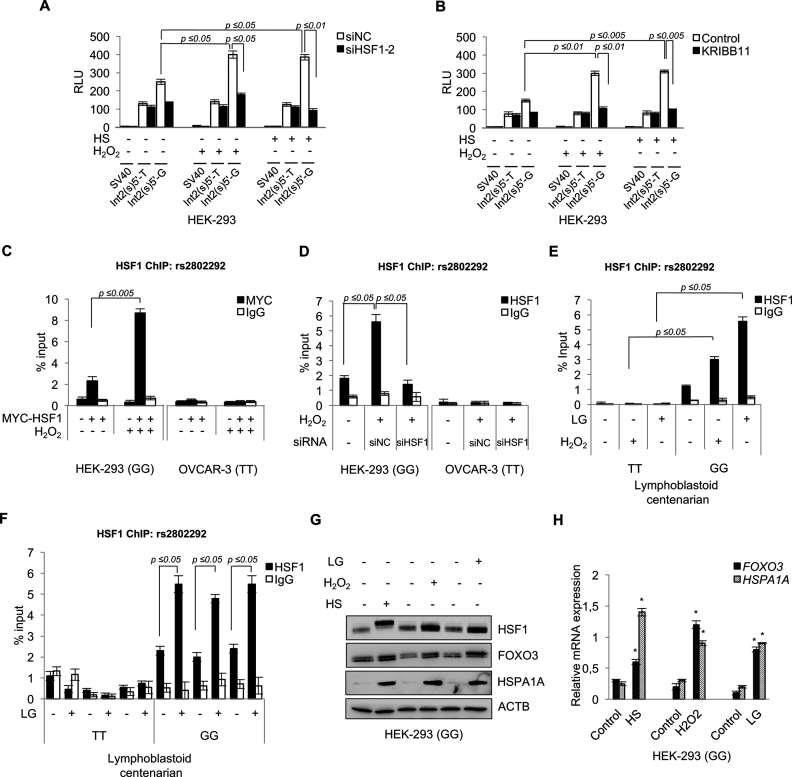
Figure 2.
HSF1 binds to the genomic rs2802292 SNP region in cells homozygous for the rs2802292 G-allele and induces the expression of FOXO3 in a stress-dependent manner. Luciferase reporter assays (n = 3) measuring the effect of HSF1 silencing by siRNA (siHSF1-2) (A) or HSF1 chemical inhibition (1 h KRIBB11, 10 μM) (B) on the enhancer activity of the rs2802292 SNP region upon hydrogen peroxide (1 h H2O2, 100 μM) or heat shock (HS) (2 h at 43°C) treatment. Cells (HEK-293) were collected and analyzed after 2 h of recovery from the indicated treatment. (C) ChIP assays (n = 3) of HSF1 occupancy of the genomic rs2802292 SNP region in HEK-293 cells homozygous for the rs2802292 G-allele (GG) in the presence or absence of exogenous MYC-tagged-HSF1 with and without H2O2 treatment (1 h, 100 μM). (D) ChIP assays (n = 3) testing the effect of HSF1 silencing by siRNA (siHSF1: siHSF1-2) on HSF1 binding to the genomic rs2802292 SNP region in HEK-293 cells homozygous for the rs2802292 G-allele (GG) and OVCAR-3 cells homozygous for the rs2802292 T-allele (TT) with and without H2O2 treatment (1 h, 100 μM). (E) ChIP analysis (n = 3) of HSF1 occupancy of the genomic rs2802292 SNP region in homozygous GG and TT cells originated from centenarians upon oxidative (1 h H2O2, 100 μM) and glucose restriction (LG) (10 h, 0.75 mM) stress stimuli. (F) ChIP analysis measuring the effect of LG (10 h, 0.75 mM) on HSF1 occupancy of the genomic rs2802292 SNP region in additional homozygous GG (n = 3) and TT (n = 3) cells originated from centenarians. (C–F) IgGs were used as an immunoprecipitation control. Expression levels of FOXO3, HSF1 and HSPA1A protein (G) and FOXO3 and HSPA1A transcript (H) were assessed by immunoblot and qPCR (n = 3), respectively, in HEK-293 cells homozygous for the rs2802292 G-allele subjected to stress conditions by heat shock (HS) (2 h at 43°C), hydrogen peroxide (1 h H2O2, 100 μM) and LG (24 h, 0.75 mM). P-values were derived from t-tests: *P ≤ 0.05.