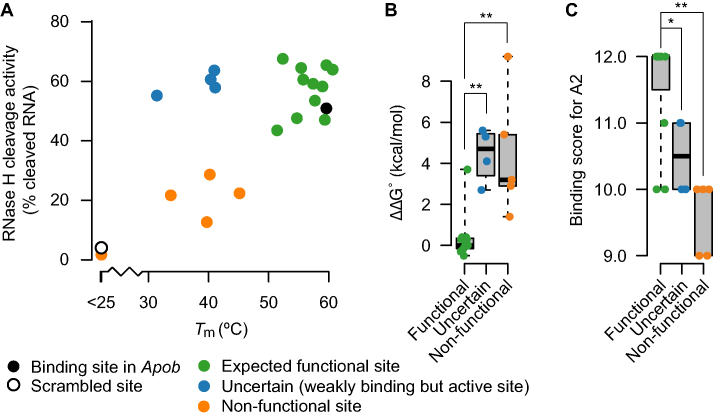
Figure 6.
Experimental characterization of potential off-target binding sites. (A) For the gapmer A2, Tm and average RNase H-cleavage activity (n = 3) toward the expected binding region sequence in 23 potential off-targets are shown. Sequences are divided into three groups based on their Tms and cleavage activities. Sequences represented by green dots had characteristics similar to the perfectly complementary Apob binding site (black dot). Sequences represented by blue dots had lower Tms, and sequences represented by orange dots had both lower Tm and lower cleavage activity, similar to the scrambled control characteristics (open dot). For the three groups of binding region sequences, differences in (B) ΔΔG values and (C) binding scores were evaluated by Wilcoxon rank sum test. *P < 0.05, **P < 0.01.