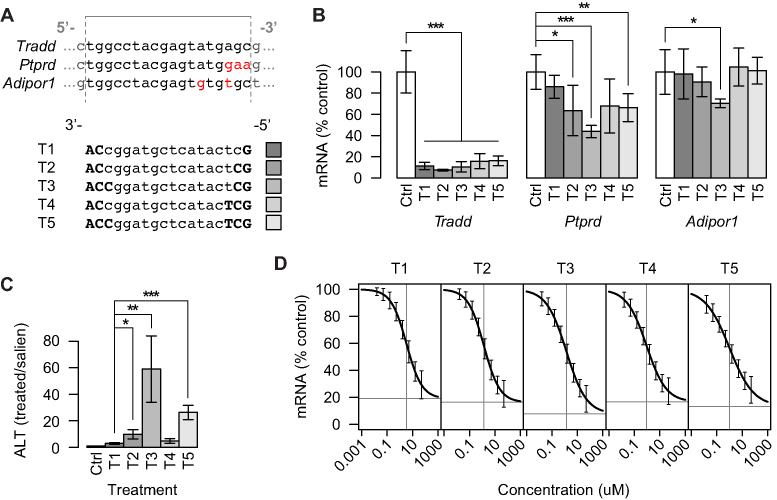
Figure 7.
Evaluation of gapmer activity on intended and unintended targets for five gapmers with the same target regions but different binding affinities. (A) Expected binding regions in Tradd (intended target) and two unintended targets Ptprd and Adipor1. Mismatched bases in the five gapmers T1–T5 are indicated in red. Gray lines indicate gapmer binding region. For gapmer sequences, uppercase bold indicates LNA and lowercase indicates DNA. (B) Knockdown of the intended target, Tradd mRNA and two unintended targets, Ptprd and Adipor1 mRNA, in mouse liver following five 15 mg/kg doses of gapmers T1–T5 over 2 weeks as measured by qRT-PCR. (C) Levels of ALT in mouse serum following five 15 mg/kg doses of gapmers T1–T5 over 2 weeks. Data represent the average ± one standard deviation relative to saline-treated controls (Ctrl) with n = 5 per treatment group. Significance of differences in average transcript levels between groups were evaluated by Student’s t-test. *P < 0.05, ***P < 0.001. (D) Knockdown of Tradd mRNA at eight different concentrations in mouse primary hepatocytes for each of the gapmers T1–T5. CRCs found by least squares fitting of the four-parameter logistic function. Error bars indicate model-based standard errors (n = 4). Horizontal and vertical gray lines indicate estimated maximal efficacy and EC50, respectively.