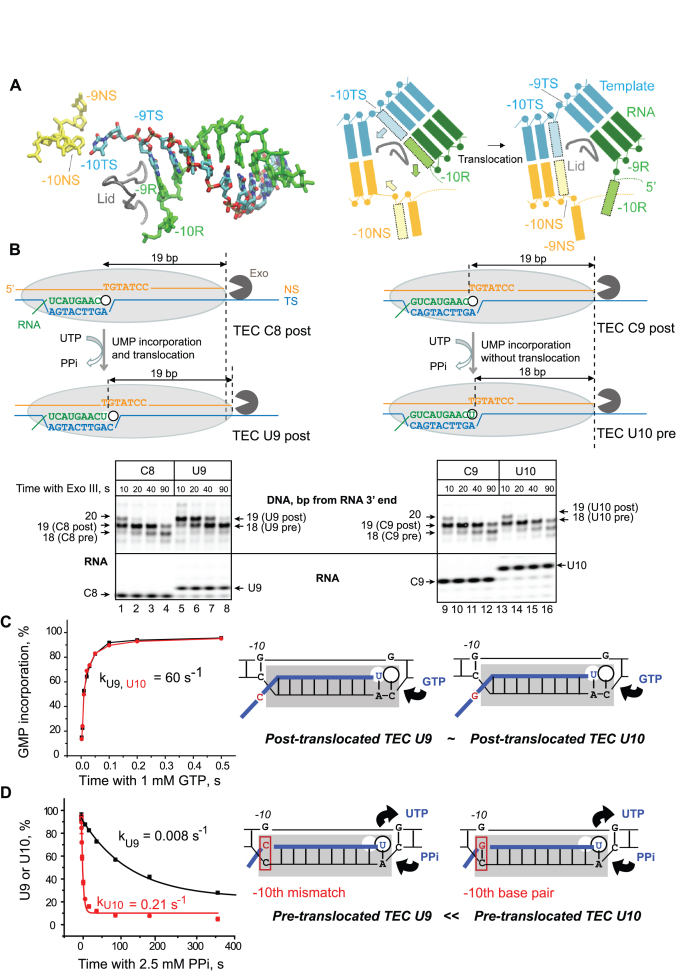
Figure 1.
RNA–DNA-10 base pairing at the upstream end of the RNA–DNA hybrid in RNAP affects pyrophosphorolysis and translocation, but not NMP addition in a non-paused TEC. (A) Cryo-EM structure (PDB: 6ALG) of the rear end of transcription bubble in the post-translocated TEC by E. coli RNAP arrested by Nun protein (8). The non-transcribed DNA strand (NS, yellow), template strand (TS, cyan), and nascent RNA (R, green) in the RNA–DNA hybrid are shown. The lid domain in the β’ subunit of RNAP (249–265 aa) is shown as a tube model (gray). The cartoon on the right illustrates RNA–DNA rearrangement at the upstream edge of the bubble during forward 1-bp translocation of RNAP (Pre→Post transition). Melting of the –10th RNA–DNA base pair in the hybrid is coupled with re-annealing of the –10th DNA–DNA base pair at the rear end of the bubble are shown as shaded boxes with the arrows indicating movement of the corresponding bases. (B) Exonuclease mapping of the front boundary of RNAP. The C8 and C9 TECs were assembled with RNA C8 or RNA C9 hybridized to TDS57 carrying three phosphothioate bonds at the 3′ end, and NDS57 labeled at the 5′ end by Cy3 (Supplementary Figure S1). The RNAP footprint is shown by a light grey oval. Two possible outcomes, UMP incorporation followed by translocation and UMP incorporation without translocation, are illustrated. The TECs were incubated with Exo III for 10, 20, 40 and 90 s before (C8 and C9) or after (U9 and U10) addition of 10 μM UTP. The RNA was detected in the same gel as the DNA to confirm UMP incorporation. GMP incorporation (C) and pyrophosphorolysis (D) were analyzed in TECs U9 and U10. The cartoons depict the apparent relative abundance of the pre- and post-translocated states in the TECs U9 and U10.