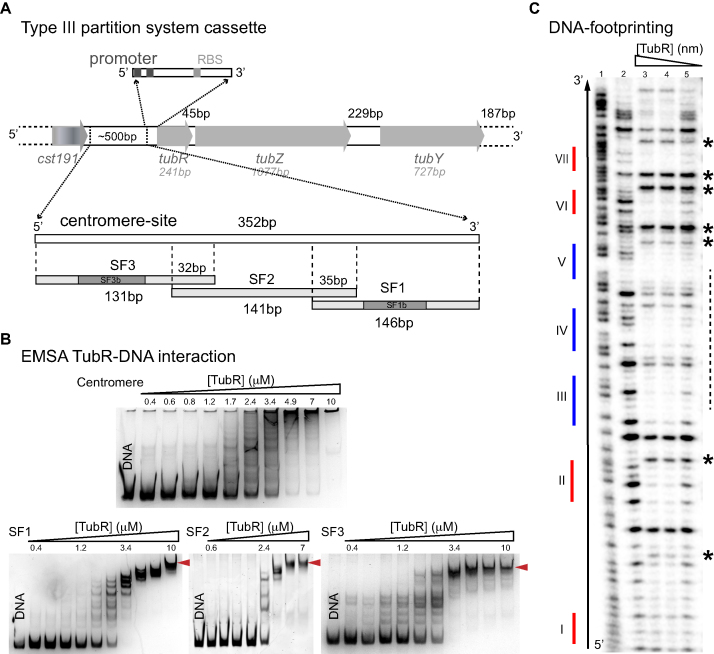
Figure 1.
tubC characterization and interaction with TubR. (A) Scheme of c-st encoded Type III partition systems including the number of bp of each gene and intergenic regions, and details related to the centromere-site and the promoter regions. The 352 bp tubC site was split into three overlapping fragments named SF1, SF2 and SF3. Fragments SF1 and SF3 show the localization of derived fragments SF1b and SF3b (related to Figure 2A). (B) EMSA experiments of 0.2 μM of each DNA fragment and increasing TubR concentrations in the presence of 0.5 mg/ml heparine as DNA competitor. EMSAs show the formation of multiple complexes that converge into a single one (red arrows) at protein saturating concentrations. Protein concentrations are 0.4–10 μM for all DNA fragments except for SF2 (0.6–7 μM). (C) DNase I footprinting experiments of 4 nM radiolabeled SF1 forward strand. Sequence of free DNA is shown by Maxam reaction (lane 1), free DNA (lane 2), 852 nM TubR (lane 3), 400 nM TubR (lane 4) and 252 nM TubR (lane 5) of TubR. Red and blue color lines represent the position of each iteron within the fragment (numbering is related to Figure 2A). Over-exposed bands (asterisks) and primary binding region in forward strand (dashed line) are marked on the right of the gel.