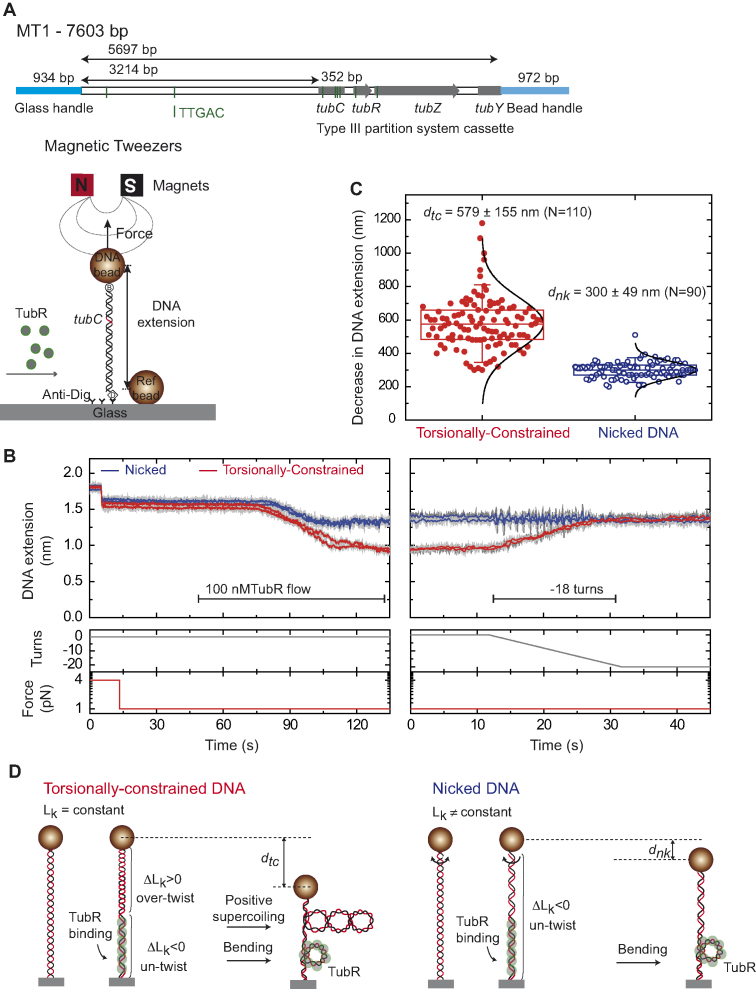
Figure 5.
TubR bends and untwists the DNA upon binding. (A) Schematic representation of the MT1 molecule used in MT experiments where the DNA substrate contains the c-st partition operon, and the experimental configuration used to measure TubR binding with magnetic tweezers. (B) TubR binding assay. DNA molecules were first identified as tcDNA or nicked by doing a rotation curve (Supplementary Figure S7). At 1 pN stretching force, 100 nM TubR was injected in the fluid cell while the extension of the DNA molecules was recorded. Different amplitudes of extension reduction were observed for tcDNA and nicked DNA. The extension of tcDNA molecules increased upon negative (clockwise) rotations of the magnets (right panel). (C) Histogram of extension reduction for tcDNA and nicked DNA molecules upon TubR incubation. (D) Cartoons depicting the interpretation of extension reduction for tcDNA and nicked DNA. TubR untwist the DNA generating positive supercoiling in tcDNA molecules (left). Nicked molecules cannot be supercoiled and therefore the reduction in extension can only be explained by bending of the DNA by TubR (right).