Abstract
In Drosophila, the most widely used system for generating spatially restricted transgene expression is based on the yeast GAL4 protein and its target upstream activating sequence (UAS). To permit temporal as well as spatial control over UAS-transgene expression, we have explored the use of a conditional RU486-dependent GAL4 protein (GeneSwitch) in Drosophila. By using cloned promoter fragments of the embryonic lethal abnormal vision gene or the myosin heavy chain gene, we have expressed GeneSwitch specifically in neurons or muscles and show that its transcriptional activity within the target tissues depends on the presence of the activator RU486 (mifepristone). We used available UAS-reporter lines to demonstrate RU486-dependent tissue-specific transgene expression in larvae. Reporter protein expression could be detected 5 h after systemic application of RU486 by either feeding or “larval bathing.” Transgene expression levels were dose-dependent on RU486 concentration in larval food, with low background expression in the absence of RU486. By using genetically altered ion channels as reporters, we were able to change the physiological properties of larval bodywall muscles in an RU486-dependent fashion. We demonstrate here the applicability of GeneSwitch for conditional tissue-specific expression in Drosophila, and we provide tools to control pre- and postsynaptic expression of transgenes at the larval neuromuscular junction during postembryonic life.
Tissue-specific expression of transgenes is a powerful tool for molecular genetic studies in model organisms. For the fruit fly Drosophila melanogaster, one of the most versatile methods to express transgenes in a spatially restricted manner is a bipartite expression system based on the yeast GAL4 protein and its upstream activating sequence (UAS; ref. 1). Since its first description, a wide variety of driver lines with defined expression patterns of the GAL4 protein have been generated, either by using cloned promoter fragments (e.g., ref. 2) or from enhancer detector screens (e.g., refs. 1 and 3). Hand in hand with GAL4 drivers, a large number of UAS reporter lines have been described (for reviews, see refs. 4 and 5). The GAL4/UAS system has a wide variety of experimental applications, such as “mosaic” expression of transgenes for cell-specific rescue of mutant phenotypes (e.g., ref. 6), but also cell ablation by using programmed cell death mechanisms (7), functional suppression of electrical excitability (35) or synaptic activity (8), as well as genetic perturbation of cellular signaling pathways (2) or cell–cell communication (9, 10). In many cases, a bipartite expression system is indispensable, especially when expression of the transgene induces lethality, hindering maintenance of the transgenic line.
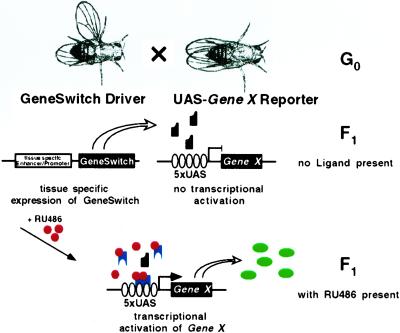
Because early dominant effects of mis- or overexpressed transgenes can preclude genetic analysis at later developmental stages or in adult animals, temporal control over the tissue-specific expression of transgenes is highly desirable. Temporal control of transgene expression in Drosophila has generally been accomplished by using a heat shock (hs) promoter. However, this approach lacks the spatial control that tissue-specific promoters provide. To circumvent this problem, several alternate approaches have been proposed. For instance, cell-specific activation of hs transgenes was achieved with a laser microbeam (11), and light-induced activation of caged GAL4 was used to activate UAS transgenes in individual embryonic cells (12). However, these approaches are technically demanding and are restricted to certain developmental stages. More recently, temporal control over tissue-specific expression in Drosophila has been accomplished by using the tetracycline-dependent “tet-off” (13, 14) or “tet-on” (15) transactivation systems, which are based on binding of tetracycline operator (tetO) sequences, either by the tetracycline repressor protein (tetR) in the absence, or by a mutant tetracycline repressor (rTA) in the presence, of tetracycline or derivatives (16, 17). These techniques are conceptually elegant but have the practical limitation in Drosophila that new tissue-specific tetR or rTA drivers as well as tetO responder lines have to be generated, although a technique linking the tet-off system to existing GAL4 drivers has recently been developed (14). We wanted to make an inducible system that could take advantage of already characterized fly lines from the GAL4/UAS system, for instance the 2,300 lines from the “EP-collection” (18, 19). Recently, modified forms of the GAL4 protein have been generated for which transcriptional activity depends on the presence of mammalian steroid hormones or chemically related compounds (20, 21). These conditional GAL4 proteins, expressed by using tissue-specific promoters and activated by systemic application of the ligand, would allow the expression of UAS transgenes in both a temporally and spatially restricted fashion. The successful application of an estrogen-receptor-GAL4 fusion protein (GAL4-ER) for temporally controlled ablation of specific cells in Drosophila oocytes has recently been reported (22). Here, we show that an RU486-dependent GAL4-progesterone-receptor fusion protein called “GeneSwitch” (21, 23) can be used for a bipartite expression system in Drosophila for spatially and temporally restricted transgene expression (Fig. 1).
Figure 1.
The GeneSwitch/UAS expression system in Drosophila. Driver lines expressing the transcriptional activator GeneSwitch in a tissue-specific fashion are crossed to UAS-reporter lines with genomic inserts of a target gene fused to five GAL4-binding sites arrayed in tandem (5× UAS). In the absence of an activator, the GeneSwitch protein is expressed in target tissues but remains transcriptionally silent (black); Gene X is therefore not expressed. However, after systemic application of RU486 (red), the GeneSwitch protein becomes transcriptionally active (blue), mediating expression of gene X (green) in only those tissues expressing GeneSwitch. [Reproduced with permission from ref. 1 (Copyright 1993, The Company of Bioligists Limited).]
Our motivation to develop an inducible system was to express transgenes during postembryonic development at the larval neuromuscular junction (NMJ). We therefore chose to use cloned promoters from two genes with either panneural (embryonic lethal abnormal vision, ELAV), or panmuscular (myosin heavy chain, MHC) distribution, to generate fly lines that express GeneSwitch, either pre- or postsynaptically at NMJs. We show that these lines can be used to express a variety of UAS transgenes in larvae in an RU486-dependent fashion. In a companion paper (24), the authors choose an enhancer detector approach to generate fly lines that express GeneSwitch in different cell types in the adult head.
We show that reporter protein expression can be adjusted by the dosage of the GeneSwitch activator, RU486, which, on its own at the concentrations used, has no overt effects on viability or fertility of flies. By using sensitive assays based on presynaptic expression of neurotoxins or genetic alteration of the physiological properties of muscles, we show very low background transcription in the absence of the activator during most of larval life. The strains expressing panneuronal or panmuscular GeneSwitch are thus suitable for temporally restricted expression either pre- or postsynaptically of UAS transgenes at the larval NMJ. More generally, we anticipate that the GeneSwitch system will be useful in other cases where temporal control over spatially restricted UAS transgene expression in Drosophila is of crucial importance.
Materials and Methods
Molecular Biology and Transformation of Drosophila.
For details of the construction of pP{ELAV-GeneSwitch} and pP{MHC-GeneSwitch}, see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org. The transformation vectors were partially sequenced to confirm correct transition sites. Transformation vectors and the helper vector pπ25.7wcΔ2–3 were grown in the Escherichia coli strain DH10b (Life Technologies, Rockville, MD) and purified by using the Qiagen MaxiPrep Kit (Qiagen, Valencia, CA). Standard procedures for P-Element-mediated transformation of Drosophila were used (25). A single homozygous viable line was generated with P{ELAV-GeneSwitch} on the third chromosome, and five independent homozygous viable lines were obtained with P{MHC-GeneSwitch}, also on the third chromosome.
Fly Stocks.
All stocks were in a yw or w1118 background and were cultured on cornmeal–molasses medium at 25°C or room temperature. ELAV-GAL4 was used for constitutive expression of UAS-transgenes in the nervous system (2). UAS lines used were UAS-TNTE, which encodes the tetanus toxin light chain (TeTxLC; ref. 8); UAS-2× enhanced green fluorescent protein (EGFP), a dicistronic EGFP (Marc S. Halfon and H.K., unpublished work); and UAS-EKO (35), a GFP-tagged noninactivating Shaker K+ channel protein. Both UAS-TNTE and UAS-EKO are embryonic lethal when panneuronally expressed by using ELAV-GAL4 (8, 35) but are viable to third instar when expressed in muscles by using 24B-GAL4.
RU486 Induction Protocols and Viability Assays.
Larvae were raised on apple juice plates or in standard fly food to the desired age. Up to 10 mg/ml of RU486 (Mifepristone, Sigma) was dissolved in ethanol or DMSO. For larval feeding, RU486 was diluted 25-fold from the working concentration in ethanol and directly mixed with the larval food to get a final concentration of 4% ethanol. For “larval bathing,” third instar larvae were washed with water and ethanol and transferred to 0.3-ml 3-mg/ml RU486 in a 1.5-ml sample tube, where they resided for 2 min before they were aged for 0–21 h on a well-yeasted apple juice plate. To activate GeneSwitch in embryos, we fed the mothers yeast paste containing RU486 for 2 days before egg collection. Embryonic or adult viability was expressed as the percentage of embryos hatching or the percentage of first instar larvae surviving to adulthood.
Protein Chemistry.
For quantitative Western blots, larvae were washed and frozen in liquid nitrogen. Crude protein extracts were made by homogenizing larvae in protein extraction buffer (1.5% SDS/Tris⋅HCl, pH 6.8). Total protein from one larva per lane was subjected to SDS/PAGE (26) and electrotransferred (27) to nitrocellulose membranes (Schleicher & Schuell). Immunodetection of GFP was performed by using a mAb against GFP (CLONTECH) and the SuperSignal ECL detection Kit (Pierce) according to the manufacturer's recommendations. Western blot data were quantified by using the Ultraviolet Products Epi Chemi II Darkroom and the labworks image analysis software package, Ver. 4.0 (Ultraviolet Products). GFP levels were normalized to levels of animals expressing GFP from a constitutively active ELAV-GAL4 driver.
Immunohistochemistry and Confocal Microscopy.
Filleted larval bodywall preparations were done as in ref. 28. Briefly, fillets were fixed in 4% paraformaldehyde for 40 min before blocking with 1% BSA and successive incubations with anti-even-skipped mAb 3C10 and AlexaFluor 568-labeled secondary Ab (Molecular Probes). Analysis of GFP expression levels was done on a Bio-Rad MRC 1024 confocal microscope by using lasersharp, Ver. 3.0 (Bio-Rad). For comparison of expression levels between preparations, identical parameters for excitation and detection were used.
Muscle Physiology.
Early third instar larvae (within 50–70% of the size of crawling third instar larvae) were filleted and pinned in Sylgard-coated slides to expose bodywall musculature. Recordings were made on ventral longitudinal muscle fiber 6 in segments A2–A4. For voltage-clamp experiments, muscles were impaled with sharp microelectrodes (15–25 MΩ, 3 M KCl) and clamped at −80 mV. Command steps ranged from −80 mV to +20 mV in 10-mV increments. Signals were amplified by using a Dagan 8500 Intracellular preamp clamp (Dagan Instruments, Minneapolis), digitized with a Digidata 1320A (Axon Instruments, Foster City, CA) and analyzed by using the pclamp 8.0 software package (Axon Instruments). For voltage-clamp recordings, a zero Ca2+ solution was used to prevent muscle contraction and contamination of IEKO by ICa or ICa/K and contained (in mM): 140 NaCl/5 KCl/0.5 EGTA/4 NaHCO3/6 MgCl2/5 N-tris[hydroxymethyl]methyl-2-aminoethanesulfonic acid/5 trehalose/50 sucrose, pH 7.2.
Results
ELAV-GeneSwitch Drives Gene Expression in Larval Neurons in an RU486-Dependent Manner.
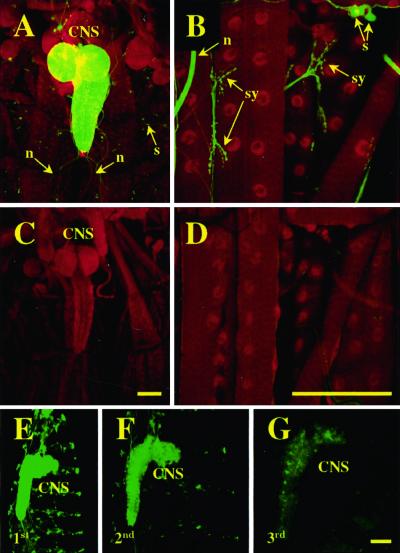
How GeneSwitch is thought to function is shown in Fig. 1. The protein is expressed in specific cells by means of a tissue-specific promoter and accumulates in nuclei in its inactive form. On binding ligand RU486, GeneSwitch is switched on as a transcriptional activator of UAS transgenes. By using UAS-2×EGFP driven by ELAV-GeneSwitch as a reporter, we investigated RU486-dependent expression of GFP in the nervous system of third instar larvae. First instar larvae homozygous for both ELAV-GeneSwitch and UAS-2×EGFP (2× EGFP; ELAV-GAL4) were transferred to food containing 15 μg/ml of RU486 or solvent only as controls. Third instar larvae were dissected, fixed, and stained with an antibody for the even-skipped protein as a nuclear marker for muscles and neurons. Confocal imaging demonstrated panneuronal GFP expression in animals raised on RU486 (Fig. 2 A and B) but essentially none in control animals (Fig. 2 C and D). In induced animals, cytosolic GFP expression shows the same distribution observed with other ELAV-based GAL4 drivers, with high levels of expression throughout the larval central nervous system (Fig. 2A) and in peripheral sensory neurons as well as at all NMJs. Thus, in these animals, RU486 mediates reporter gene expression in the expected pattern of the ELAV-GeneSwitch driver.
Figure 2.
RU486 activates GeneSwitch in the larval nervous system. (A–D) Confocal images of fixed bodywall preparations of Drosophila third instar larvae expressing cytosolic GFP (green channel) from a neuron-specific GeneSwitch driver. Larvae were raised in the presence (A, B) or absence (C, D) of RU486. Counterlabeling is for the nuclear localized even skipped (red channel). (E–G) Confocal images of unfixed, whole-mount first (E), second (F), or third (G) instar larvae expressing UAS-EGFP from ELAV-GeneSwitch. GeneSwitch was activated in embryos by feeding mothers RU486, but first instar larvae were transferred to, and developed on, normal food. (CNS, central nervous system; s, sensory neurons; n, nerves; sy, neuromuscular synapses.) (Bar = 100 μm.)
We further examined the effects of removing RU486 on GFP reporter gene expression driven by ELAV-GeneSwitch. We fed mothers RU486 for 1–2 days before egg collection and examined GFP fluorescence in the progeny. Newly hatched first instar larvae expressed GFP in the nervous system (Fig. 2E), indicating transfer of RU486 from the mothers to the embryos and activation of the embryonic ELAV-GeneSwitch. If these larvae were raised in the absence of RU486, GFP fluorescence in neurons markedly decreased after 24 h (Fig. 2F) and largely disappeared by third instar (Fig. 2G). Thus, the temporal activity of GeneSwitch can be regulated by the presence or absence of RU486 in the larval food.
GeneSwitch Mediates Transcription in a Time- and RU486 Dose-Dependent Fashion.
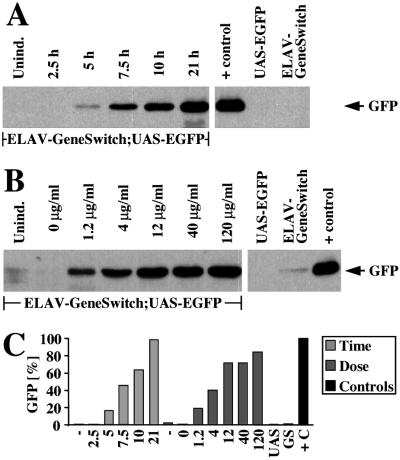
To address the kinetics and dose dependence of protein expression in response to RU486, we quantified induced GFP levels after RU486 application to third instar larvae by means of Western blot, immunodetection, and densitometry. GFP was detectable 5 h after application of RU486 by “larval bathing” (16.2% of positive control level, Fig. 3 A and C). Robust expression was seen after 7.5 h, and by 21 h, reporter protein levels were almost identical (98.4%) to GFP detected in a fly strain with GFP expressed from constitutive GAL4 under the control of the ELAV promoter. Similarly, GFP was detectable in third instar larvae reared on food containing as little as 1.2 μg/ml of RU486 (19.0%, Fig. 3 B and C). Reporter protein expression showed dose dependence up to 12 μg/ml of RU486 (71.7%) but did not substantially increased by up to 10-fold higher concentrations of RU486 (84.4% at 120 μg/ml). Hence, by feeding 12–120 μg/ml of RU486, a 51- to 60-fold increase in reporter protein expression could be achieved relative to basal GFP protein levels in the absence of RU486 (1.4 ± 1.1%).
Figure 3.
RU486-mediated expression starts rapidly and is dose-dependent. Quantitative Western blot analysis of GFP protein. Genotypes of the larvae are ELAV-GeneSwitch; UAS-EGFP if not otherwise indicated. UAS-EGFP;+ (UAS) and +;ELAV-GeneSwitch (GS) are parental lines; ELAV-GAL4;UAS-EGFP animals have constitutively active GAL4 in all neurons serve as a positive control (+ control, +C). (A) Timecourse of GFP protein (arrow) expression (hours after “larval bathing”) and (B) dose dependence of GFP protein expression on RU486 concentration (μg/ml) in larval food. (C) Qantitative analysis of Western blots in A (light gray, “Time”) and B (dark gray, “Dose”) normalized to positive control (ELAV-GAL4;UAS-EGFP, black).
ELAV-GeneSwitch Shows Very Low Transcriptional Activity in the Absence of RU486.
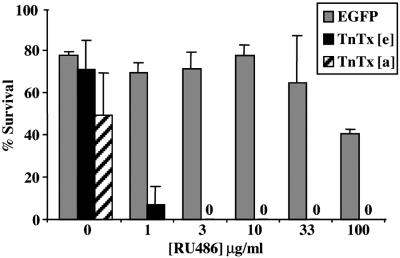
Both fluorescence and Western blot analysis suggested that the basal transcriptional activity of GeneSwitch in the absence of RU486 was very low in vivo. As a stringent test of “leakiness,” we examined nervous system expression of TeTxLC, scoring for lethality. TeTxLC is one of the most potent of the Clostridium neurotoxins and blocks chemical synaptic transmission at femtomolar concentrations (29). Panneural expression of TeTxLC (by using the UAS-TNTE transgene driven by ELAV-GAL4) causes complete embryonic paralysis and failure to hatch (8, 35). We generated embryos with a UAS-TNTE transgene driven by the panneuronally expressed ELAV-GeneSwitch. In the absence of RU486, embryonic survival of UAS-TNTE animals expressing GeneSwitch (Fig. 4, black columns) was not significantly different from control animals with UAS-EGFP (gray columns, 71.5 ± 13.9 vs. 77.8 ± 1.5%). However, when ELAV-GeneSwitch in the embryo was activated by feeding the parents yeast with RU486 before the experiment, embryonic survival dropped dramatically to almost zero (7.0 ± 8.5% for 1 μg/ml of RU486, 0% survivors for 3 μg/ml of RU486 or higher concentrations). RU486 treatment of control animals (gray columns) had no significant effect on embryonic viability up to 33 μg/ml in parental food. Larvae expressing ELAV-GeneSwitch and carrying the UAS-TNTE transgene showed decreased viability, even in the absence of RU486. In several independent experiments, 36–72% of ELAV-GeneSwitch/UAS-TNTE first instar larvae survived to adulthood (49 ± 20%, n = three experiments with 30–50 first instar larvae each). The reduced viability of uninduced ELAV-GeneSwitch/UAS-TNTE animals indicates that there is low but nonetheless significant basal expression of the transgene at postembryonic stages.
Figure 4.
Both basal transcriptional activity of ELAV-GeneSwitch and toxicity of RU486 are low. The viability of embryos expressing either a UAS-TNTE reporter (black bars, TnTx[e]) or a control UAS-EGFP reporter (gray bars, EGFP) from the ELAV-GeneSwitch driver is shown as a function of RU486 concentrations in the parents' food. The percentage of UAS-TNTE/ELAV-GeneSwitch first instar larvae surviving to adulthood in the absence of RU486 is shown as a hatched bar (TnTx[a]).
MHC-GeneSwitch Confers RU486-Dependent Transgene Expression in Larval Muscles.
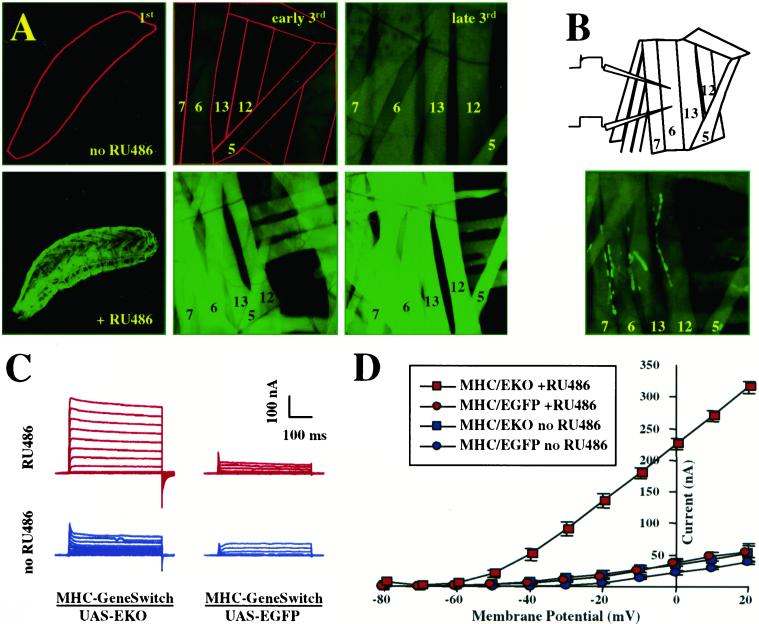
To examine the efficacy of GeneSwitch when expressed in larval muscles, we constructed transgenic flies with GeneSwitch under the control of the MHC promoter (MHC-GeneSwitch). Larvae carrying both UAS-EGFP and MHC-GeneSwitch transgenes showed essentially no GFP fluorescence up to early third instar in the absence of RU486 (Fig. 5A Top Left and Middle), whereas their siblings raised in the presence of 50 μg/ml of RU486 show strong GFP expression in bodywall muscles (Fig. 5A Bottom Left and Middle). In contrast to the low basal transcriptional activity of GeneSwitch without RU486 in the nervous system, GFP fluorescence unexpectedly becomes visible during the late third larval instar in uninduced animals (Fig. 5A Top Right).
Figure 5.
RU486 induces transgene expression in larval bodywall muscles. (A) Expression of UAS-EGFP driven by MHC-GeneSwitch in bodywall muscles of first (Left), early third (Center), and late third instar larvae (Right) in the presence (Bottom) or absence (Top) of RU486. The bodywall musculature was imaged through the cuticle in undissected larvae. Muscle fibers 7, 6, 13, 12, and 5 are labeled; first instar larva or muscles are outlined in red in uninduced animals for better visibility. (B) Schematic diagram for two electrode voltage-clamp analyses (Upper) and EKO-channel localization predominantly to NMJs in fixed bodywall preparations from third instar UAS-EKO/MHC-GeneSwitch larvae (Lower). (C) Representative current traces and (D) current–voltage relationship obtained under zero Ca2+ conditions for uninduced (blue traces and curves) or induced (red traces and curves) UAS-EKO/MHC-GeneSwitch animals (left column in C, squares in D) or UAS-EGFP/MHC-GeneSwitch controls (right column in C, circles in D).
GeneSwitch-Mediated Expression of Ion Channels Can Be Used to Manipulate Neuromuscular Activity in an RU486-Dependent Fashion.
To estimate the relative levels of gene expression in the presence and absence of RU486, we used UAS-EKO as a reporter. The UAS-EKO transgene consists of a noninactivating form of the Shaker K+ channel, optimized to serve as a current shunt and vitally tagged with EGFP (the EKO-channel), which can be expressed from the GAL4 UAS. EKO channels expressed in larval bodywall muscles are localized by discs-large to the postsynaptic membranes and elevate K+ conductance in a dosage-dependent fashion (35). Thus, K+ conductance of larval bodywall muscles expressing the EKO-transgene can be used as a quantitative readout of EKO-channel expression levels.
We generated larvae expressing UAS-EKO from the MHC-GeneSwitch driver and raised them in the presence of 50 μg/ml of RU486 to activate EKO expression. By early third instar, the GFP-tagged EKO channel is expressed in induced larvae in all of the bodywall muscles and predominantly localizes, as expected, to the NMJs (Fig. 5B). We measured K+ currents by two-electrode voltage-clamp analysis on MF6 in abdominal segments 2–4 (for a schematic representation, see Fig. 5B). In RU486-induced animals, we saw outward K+ currents of 315 ± 9 nA at a holding potential of +20 mV, which is a 7-fold increase over induced control animals (MHC-GeneSwitch/UAS-EGFP; 55 ± 10 nA at +20 mV; see Fig. 5 C and D, red traces). In uninduced animals, peak K+ currents at +20 mV reached only 53 ± 15 nA in the presence of EKO channels, a 36% increase over the 39 ± 3 nA measured in uninduced controls (Fig. 5 C and D, blue traces). Therefore, RU486-induced EKO expression in muscles led to an increase in K+ currents 20-fold higher (708 vs. 36%) than basal, uninduced expression.
Discussion
A method for temporally and spatially restricted expression of transgenes would be an invaluable tool for mosaic analysis in model organisms. In Drosophila, the most widely used method for spatially restricted expression is based on a bipartite system in which the yeast GAL4 protein controls expression of UAS transgenes (1, 4, 5). This makes a conditional form of the GAL4 protein a favorable tool for applications in Drosophila, because it is compatible with a large variety of existing genetic tools.
RU486-Activated GeneSwitch Mediates Protein Expression in Vivo in Drosophila.
We demonstrate here that the conditional “GeneSwitch” GAL4 protein (21, 23) can be used to induce expression of UAS reporters in vivo in Drosophila. The GeneSwitch protein can be expressed in a mosaic fashion by using specific promoters and is activated by adding the drug RU486 (mifepristone) to the animal's diet (Fig. 2 A and B). Notably, RU486 is transferred from mothers into embryos (Fig. 2E), and induced expression by using GeneSwitch is indistinguishable from expression mediated by constitutive GAL4 protein under the same promoters in tissue types as distinct as muscles (Fig. 5A) and neurons (Fig. 2). We conclude that RU486 passes cellular barriers, such as the larval blood–brain barrier (30), and readily penetrates the nucleus of these cells, and we expect that the same will hold true for other Drosophila cell types.
At doses used for activation, RU486 alone was not observed to be toxic for larvae or adults. Adults can tolerate dosages as high as 215 μg/ml without apparent deleterious effects (24). Similarly, the viability of embryos was unaffected by feeding mothers doses up to 33 μg/ml of RU486 (Fig. 4). However, a higher dose of RU486 (100 μg/ml) reduces embryonic viability to ≈50%. It is possible that physiological sensitivity to RU486 is at its highest during embryonic and larval development.
Systemic application of RU486 leads to activation of UAS reporters within 5 h (Fig. 3A). Once activated, GeneSwitch-mediated transgene expression can be turned off by shifting animals to RU486-free food. The slower “off-response” (Fig. 2 E–G) may in part be due to perdurance of the highly stable GFP (31). Still, RU486-mediated activation of gene expression, as well as interruption of transgene expression by discontinuing the feeding of the drug, could prove useful for developmental studies. For instance, the maternal activation of GeneSwitch could be used to rescue embryonic lethality in mutant backgrounds and would uncover the role of the mutant gene at later stages. Likewise, dominant–negative constructs (e.g., ref. 2) or expressible double-stranded RNA constructs (32) could be used to selectively interfere with gene function at larval, pupal, or adult stages.
Because the GeneSwitch protein shows very low transcriptional activity in the absence RU486 (21), we did not expect high basal activity of the GeneSwitch system. Indeed, in embryos and during most of the larval stages, we found only very low expression of reporter genes in the absence of RU486, as demonstrated by expressing TeTxLC, a potent neurotoxin (8), in the nervous system under the control of ELAV-GeneSwitch. Although a constitutive ELAV-GAL4 or induced ELAV-GeneSwitch in trans to UAS-TNTE causes essentially complete late embryonic lethality (Fig. 4 and ref. 35), uninduced GeneSwitch allowed almost normal embryonic hatching. However, only half of the first instar larvae expressing TeTxLC under the control of uninduced ELAV-GeneSwitch survived to adulthood. Because it has previously been shown that tetanus toxin acts in vivo at femtomolar concentrations (29), we suspect that low basal activity of uninduced GeneSwitch can lead to accumulation of TeTxLC in the central nervous system to reach those levels over long time scales.
One prominent exception to the low basal activity of GeneSwitch was MHC-GeneSwitch-mediated expression in the absence of RU486 in muscles during late third instar (Fig. 5A). Interestingly, this stage is coincident with a large surge of the flies' major steroid hormone, 20-HE (33), raising the question of whether 20-HE activates GeneSwitch. A direct interaction, however, seems unlikely, because a similar surge in background expression was not observed for GeneSwitch in the nervous system (Fig. 2); However, a muscle-specific metabolite of 20-HE activating GeneSwitch cannot be ruled out. Alternatively, this phenomenon could be explained by activity of uninduced GeneSwitch at very high concentrations. High promoter activity and GeneSwitch mRNA stability conferred by nontranslated sequences in the P{MHC-GeneSwitch} construct (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org) may lead to particularly high levels of the GeneSwitch protein in muscles.
Two GeneSwitch Constructs Allow Temporally Controlled Expression at Larval NMJs.
One advantage of a temporally and spatially restricted expression system is that it allows independent genetic manipulation of interacting sets of cells at different points in development. For example, neurons and muscles interact to elaborate synapses through which they communicate. From the first contact at midembryogenesis to the fully established NMJ in third instar larvae, a variety of pre- and/or postsynaptically expressed factors orchestrate different aspects of synaptogenesis (34). By using a neuron- and a muscle-specific promoter, we constructed two fly lines that allow entirely pre- or postsynaptic conditional expression of UAS transgenes at the larval NMJ. These lines provide a unique toolset for temporally controlled genetic manipulations of the larval NMJ. We demonstrate that by using UAS-EKO or the UAS-TNTE, respectively, to induce a conditional loss of postsynaptic electrical excitability in larvae (Fig. 5 C and D) or RU486-dependent loss of synaptic transmission in embryos (Fig. 4). These experiments reveal the kind of novel opportunities available with a conditional GAL4 system: embryonic lethality caused by neuronal expression of a neurotoxin represents the most severe case of deleterious effects of transgenes early in development, precluding analysis of effects at later stages. By using the GeneSwitch system, early effects can be avoided, which permits the addressing of effects of transgenes in larvae, during metamorphosis, or on the adult nervous system in the background of normal development.
Advantages of the GeneSwitch System for Future Applications.
A conditional GAL4 protein for temporally controlled mosaic expression of transgenes has a practical advantage over non-GAL4/UAS-based systems in that all existing and well characterized UAS transgenes can be used. This feature is particularly important in cases where a considerable amount of work was necessary to generate and characterize the UAS lines, such as for the “EP-lines,” a collection of 2,300 fly lines in which random insertion of UAS sequences into the genome enables GAL4-mediated misexpression of downstream genes in Drosophila (18, 19). By using the ELAV-GeneSwitch driver, targeted misexpression in the nervous system during metamorphosis or in adults may be used to identify genes involved in the formation of the mature form of the nervous system or in adult behavior and to separate those effects from earlier developmental effects.
The other currently available conditional GAL4 protein, ER-GAL4 (20), has been demonstrated to work in Drosophila (22). We have chosen the GeneSwitch protein for an inducible system for its claimed low background expression, high inducibility, and lack of interference with endogenous gene expression (21). Although a direct comparison of the two conditional GAL4 systems in Drosophila is difficult because of the different promoters that have been used, the efficacy and simplicity of use appear to be comparable. Differences may include drug treatment (22) and timing of transgene expression [5–7.5 h to transgene detection, less then 1 day for maximal expression for the GeneSwitch system (Fig. 3A), as compared with 12 h and 2.5 days for ER-GAL4, as reported in ref. 22]. However, the availability of several alternative methods for temporally and spatially controlled gene expression in flies, including the tetracycline repressor protein/tetracycline operator-based system (13–15), would be invaluable for independent manipulation of multiple transgenes in the same animal.
Supplementary Material
Acknowledgments
We thank Dr. Liqun Luo (Department of Biological Sciences, Stanford University, Stanford, CA) for providing the ELAV promoter fragment and Dr. Akira Chiba (Department of Cell and Structural Biology, University of Illinois, Urbana) for making available the MHC promoter sequences. UAS-TNTE flies were kindly provided by Dr. Cahir O'Kane (Department of Genetics, University of Cambridge, Cambridge, U.K.). mAb 3C10 was obtained from the Developmental Studies Hybridoma Bank, maintained by the University of Iowa Department of Biological Sciences, Iowa City, IA. This work was supported by grants from the National Institutes of Health and the National Aeronautics and Space Administration (to H.K.).
Abbreviations
- UAS
upstream activating sequence
- NMJ
neuromuscular junction
- ELAV
embryonic lethal abnormal vision
- MHC
myosin heavy chain
- GFP
green fluorescent protein
- EGFP
enhanced GFP
- TeTxLC
tetanus toxin light chain
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 2.Luo L, Liao Y J, Jan L Y, Jan Y N. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 3.Gustafson K, Boulianne G L. Genome. 1996;39:174–182. doi: 10.1139/g96-023. [DOI] [PubMed] [Google Scholar]
- 4.Brand A H, Dormand E L. Curr Opin Neurobiol. 1995;5:572–578. doi: 10.1016/0959-4388(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 5.Phelps C B, Brand A H. Methods. 1998;14:367–379. doi: 10.1006/meth.1998.0592. [DOI] [PubMed] [Google Scholar]
- 6.Leiserson W M, Harkins E W, Keshishian H. Neuron. 2000;28:793–806. doi: 10.1016/s0896-6273(00)00154-9. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, Schnitzler A, Agapite J, Schwartz L M, Steller H, Nambu J R. Proc Natl Acad Sci USA. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney S T, Broadie K, Keane J, Niemann H, O'Kane C J. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 9.Lin D M, Fetter R D, Kopczynski C, Grenningloh G, Goodman C S. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 10.Harris R, Sabatelli L M, Seeger M A. Neuron. 1996;17:217–228. doi: 10.1016/s0896-6273(00)80154-3. [DOI] [PubMed] [Google Scholar]
- 11.Halfon M S, Kose H, Chiba A, Keshishian H. Proc Natl Acad Sci USA. 1997;94:6255–6260. doi: 10.1073/pnas.94.12.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cambridge S B, Davis R L, Minden J S. Science. 1997;277:825–828. doi: 10.1126/science.277.5327.825. [DOI] [PubMed] [Google Scholar]
- 13.Bello B, Resendez-Perez D, Gehring W J. Development (Cambridge, UK) 1998;125:2193–2202. doi: 10.1242/dev.125.12.2193. [DOI] [PubMed] [Google Scholar]
- 14.Stebbins M J, Yin J C. Gene. 2001;270:103–111. doi: 10.1016/s0378-1119(01)00447-4. [DOI] [PubMed] [Google Scholar]
- 15.Bieschke E T, Wheeler J C, Tower J. Mol Gen Genet. 1998;258:571–579. doi: 10.1007/s004380050770. [DOI] [PubMed] [Google Scholar]
- 16.Gossen M, Bonin A L, Freundlieb S, Bujard H. Curr Opin Biotechnol. 1994;5:516–520. doi: 10.1016/0958-1669(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 17.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 18.Rorth P. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin G M, Weigmann K, Milan M, Benes V, Ansorge W, Cohen S M. Development (Cambridge, UK) 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 20.Braselmann S, Graninger P, Busslinger M. Proc Natl Acad Sci USA. 1993;90:1657–1661. doi: 10.1073/pnas.90.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burcin M M, O'Malley B W, Tsai S Y. Front Biosci. 1998;3:c1–c7. doi: 10.2741/a258. [DOI] [PubMed] [Google Scholar]
- 22.Han D D, Stein D, Stevens L M. Development (Cambridge, UK) 2000;127:573–583. doi: 10.1242/dev.127.3.573. [DOI] [PubMed] [Google Scholar]
- 23.Burcin M M, Schiedner G, Kochanek S, Tsai S Y, O'Malley B W. Proc Natl Acad Sci USA. 1999;96:355–360. doi: 10.1073/pnas.96.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roman G, Endo K, Zong L, Davis R L. Proc Natl Acad Sci USA. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keshishian H, Chiba A, Chang T N, Halfon M S, Harkins E W, Jarecki J, Wang L, Anderson M, Cash S, Halpern M E, et al. J Neurobiol. 1993;24:757–787. doi: 10.1002/neu.480240606. [DOI] [PubMed] [Google Scholar]
- 29.Herreros J, Lalli G, Montecucco C, Schiavo G. J Neurochem. 2000;74:1941–1950. doi: 10.1046/j.1471-4159.2000.0741941.x. [DOI] [PubMed] [Google Scholar]
- 30.Carlson S D, Juang J L, Hilgers S L, Garment M B. Annu Rev Entomol. 2000;45:151–174. doi: 10.1146/annurev.ento.45.1.151. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang C C, Kain S R. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 32.Kennerdell J R, Carthew R W. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 33.Riddiford L M. The Embryonic Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 34.Budnik V, Gramates L S. Neuromuscular Junctions in Drosophila. 1999. [Google Scholar]
- 35.White B H, Osterwalder T P, Yoon K S, Joiner W J, Whim MD, Kaczwarek L K, Keshishian H. Neuron. 2001;31:699–711. doi: 10.1016/s0896-6273(01)00415-9. . (Academic, New York). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.