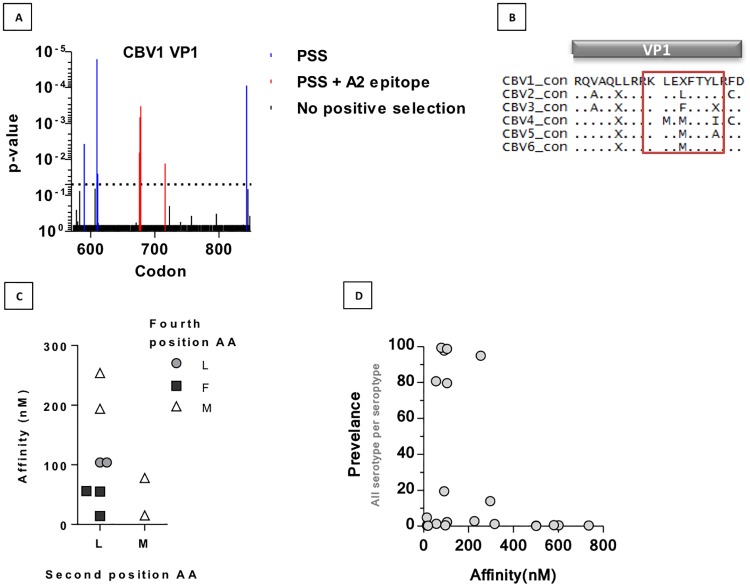
Fig 4. MEME analysis of CBV1 VP1 sequences reveal putative immunogenic regions across serotypes of CBV.
(A) Trimmed and edited CBV1 VP1 sequences from Table 4 were aligned by Bio.Align in Python and submitted to Datamonkey to identify sites of positive selection by mixed effects model of evolution. The p-value for the significance of which the amino acid site is evolving under positive selection within the model is represented. (B) Epitopes present at VP1 684–692 are highlighted within an alignment of serotype consensus sequences. (C) The predicted affinity for all potential sequences at VP1 684–692 are plotted against the amino acid identity for positions 2 and 4 in the 8-mer. (D) Prevalence of each sequence present at sites 682–694 for all serotypes was determined and plotted against their predicted binding affinity for HLA-A*0201, determined by HLA-restrictor.