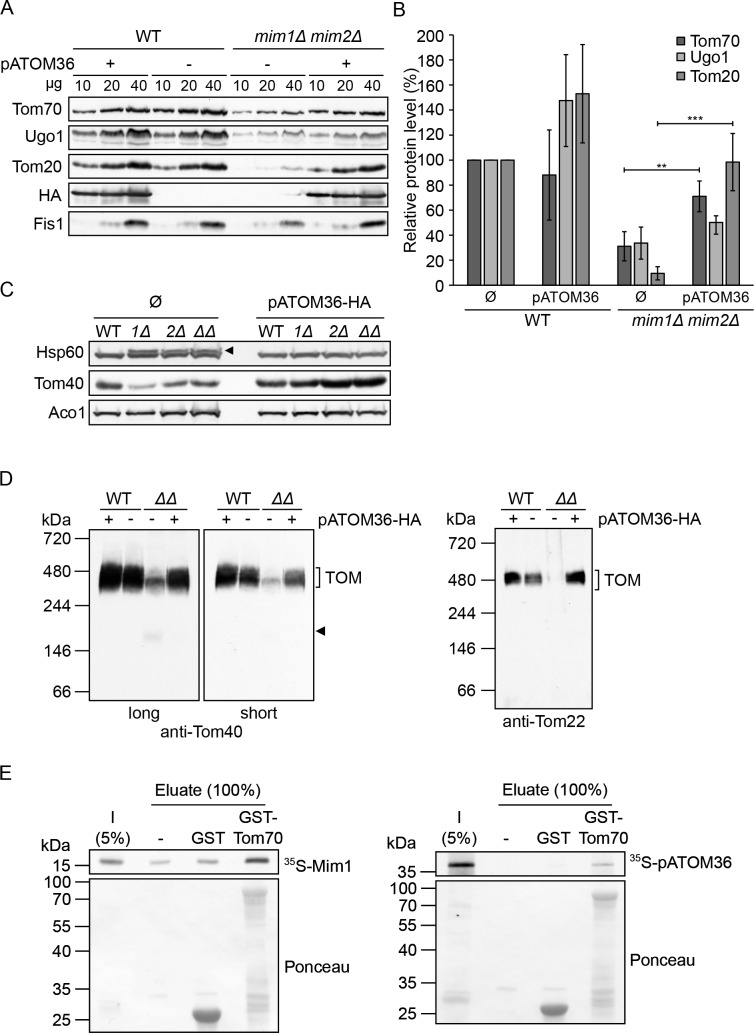
Figure 3. pATOM36 can compensate for the reduced steady state levels and assembly defects in cells lacking both Mim1 and Mim2.
(A) Mitochondria were isolated from WT or mim1Δ/mim2Δ cells transformed with either an empty plasmid (-) or with a plasmid encoding pATOM36-HA (+). The specified amounts were analysed by SDS-PAGE and immunodecoration with antibodies against either the indicated mitochondrial proteins or the HA-tag. (B) The intensity of the bands from three independent experiments such as those presented in (A) was monitored. The amounts of Tom70, Ugo1 and Tom20 in the various mitochondria samples are presented as mean percentage of their levels in control organelles (WT+ Ø). The levels of Fis1 were taken as loading control. Error bars represent ± SD. **p≤0.005, ***p≤0.0005. (C) Whole cell lysates were obtained from WT, mim1Δ (1Δ), mim2Δ (2Δ), or the double deletion mim1Δ/mim2Δ (ΔΔ) cells transformed with either an empty plasmid (Ø) or with a plasmid encoding pATOM36-HA. Samples were analysed by SDS-PAGE and immunodecoration with antibodies against the indicated mitochondrial proteins. The precursor form of mitochondrial Hsp60 is indicated with an arrowhead. (D) The mitochondria described in (A) were solubilised in a buffer containing 1% digitonin and then analysed by BN-PAGE followed by western blotting. The membranes were immunodecorated with antibodies against the TOM subunits, Tom40 (long and short exposures) and Tom22. The TOM complex is signposted. A Tom40-containing low molecular weight complex is indicated with an arrowhead. (E) Mim1 and pATOM36 interact directly with Tom70. Radiolabelled Mim1 or pATOM36 (input, I) were incubated with glutathione beads (-) or with beads that were pre-bound to recombinant GST alone or to GST fused to the cytosolic domain of Tom70 (GST-Tom70). After washing, bound material was eluated and proteins were analysed by SDS–PAGE followed by blotting onto a membrane, and detection with either autoradiography (upper panel) or Ponceau staining (lower panel).