Fig. 2.
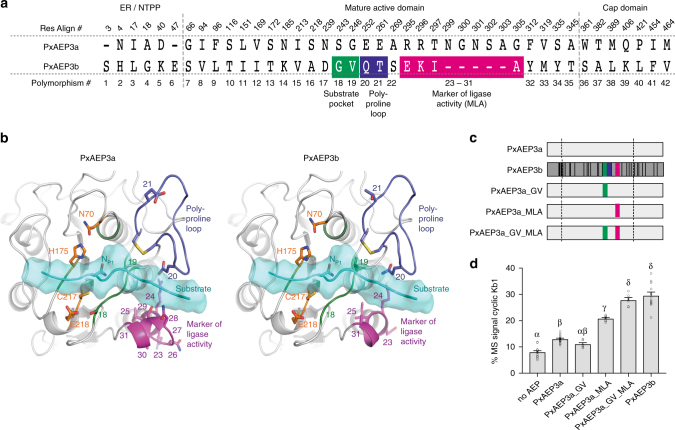
Identification and functional testing of the key structural features unique to the petunia ligase PxAEP3b. a Polymorphic residues between petunia isoforms PxAEP3a and PxAEP3b (see also Supplementary Fig. 3 for residue alignment). Those closest to the substrate pocket include polymorphisms 18 and 19 (green), polymorphisms 20 and 21 (blue) within the poly-proline loop and polymorphisms 23–31 (magenta) here named the marker of ligase activity (MLA). b Homology model of the substrate-binding site of the catalytic domain of PxAEP3a (left) and PxAEP3b (right). A substrate peptide bound in the binding site (cyan) was modeled by homology using the structure of human AEP (PDB ID: 4awa; www.rcsb.org) bound to a suicide substrate and extended at the C-terminus by a polyalanine peptide. The asparagine residue in the P1 pocket is noted NP1. Polymorphic positions in and around the substrate pocket are shown as sticks, numbered and colored according to panel a. The catalytic residues are represented in orange sticks and numbered according to homologous residues on OaAEP1b. c Schematics of petunia AEP genes prepared to test the influence of polymorphic residues on the in planta cyclization of the kB1 precursor. Colors used in the bars represent the same regions and colors in panel a. d MALDI- MS analysis of N. benthamiana-produced peptides upon co-expression of petunia AEP variants with Oak1. For all treatments, means and s.e.m. are of the percentage MS signal obtained for cyclic kB1 over the total MS signal of all kB1-related peptides produced. Treatments carrying unique Greek lettering are significantly different (P < 0.05) as determined by Tukey’s ANOVA. Replicates are as follows: n = 14 (No AEP), n = 16 (PxAEP3a), n = 6 (PxAEP3a_GV), n = 8 (PxAEP3a_MLA), n = 6 (PxAEP3a_GV-MLA), and n = 12 (PxAEP3b). Error bars are s.e.m.