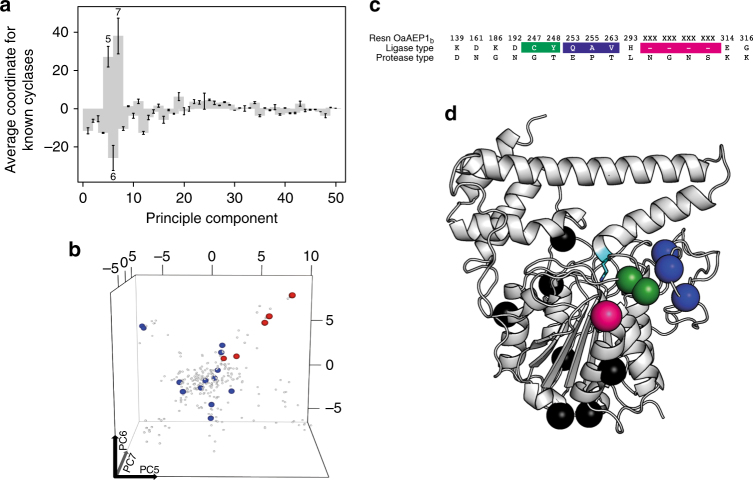
Fig. 5.
Plant AEP protein sequence space modeling. a Average protein sequence space coordinates of the six known ligases (Supplementary Data 1, Supplementary Table 2). b Plant AEP protein sequence space profiles after principle component analysis. Each point indicates a sequence, with its coordinates in the space determined by its residue properties. Axes are principle components 5, 6 and 7, which showed significant separation of known ligases (red) and proteases (blue) (Supplementary Table 2). c The 16 ligase predictive residues and positions on OaAEP1b. Color shading used is the same as in Fig. 2a. d Structure of the zymogen OaAEP1 (PDB accession 5HOI) with positions of ligase-preferred residues displayed. Residues shown in green include those closest to the substrate pocket, which includes the Gatekeeper residue Cys247 and the adjacent Tyr248. Those in blue are situated within the plant AEP-specific poly-proline loop, while the magenta colored sphere represents the position of the 4-amino acid deletion in the MLA. The seven spheres in black represent the residues most distal from the catalytic active site