Abstract
Three new cyclic peptides including a cyclic tetrapeptide (1), an aspochracin-type cyclic tripeptide sclerotiotide L (2) and a diketopiperazine dimer (3), have been isolated from the ethyl acetate extract of a marine sponge-derived fungus Aspergillus violaceofuscus. The structures of all compounds were unambiguously elucidated on the basis of HRESIMS, 1D and 2D NMR spectroscopic data, MS/MS experiments and chemical methods. Compounds 1 and 3 showed anti-inflammatory activity against IL-10 expression of the LPS-induced THP-1 cells with inhibitory rates of 84.3 and 78.1% respectively at concentration of 10 μM.
Keywords: sponge-derived fungus, Aspergillus violaceofuscus, cyclic peptides, structural characterization, anti-inflammatory
Introduction
As a class of important metabolites from marine-derived organisms, cyclic peptides are extensively present in marine tunicate (Ireland et al., 1982), sponge (Zhang et al., 2010), algae (Xu et al., 2008), bacteria (Teta et al., 2017), fungi (Bao et al., 2013), etc., and often these cyclopeptides possess rare molecular skeleton (Fukuhara et al., 2015). Moreover, due to versatile biological functions including antineoplastic (Ireland et al., 1982), antimicrobial (Teta et al., 2017), anti-inflammatory (Randazzo et al., 2001), antitubercular (Daletos et al., 2015) and histone deacetylase inhibitory activities (Gu et al., 2007), the cyclic peptides have received enduring attention of organic chemists, biologists and pharmacologists. The structures of cyclic peptides may contain unusual amino acids or be modified by methylation (Jang et al., 2017), acetylation, lipidation (Luo et al., 2014), and sulfuration (Fukuhara et al., 2015). These characteristics are playing a vital role in the interactions with relevant bioactive targets (Sieber and Marahiel, 2005; Raaijmakers et al., 2010).
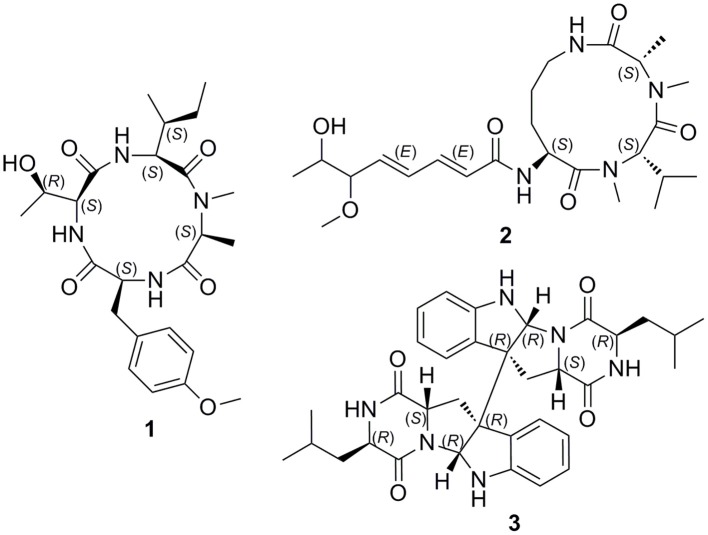
The sponge-derived fungi have been proven to be a prolific source of cyclic peptides (Amagata et al., 2006; Yu et al., 2008). In previous search for structurally unique cyclic peptides from marine sponge-derived fungus Nigrospora oryzae PF18 collected off the Xisha Islands in the South China Sea, we have identified a series of new cyclohexadepsipeptides oryzamides A–C (Ding et al., 2016). As part of our continuing quest for new bioactive molecules, chemical investigation of secondary metabolites of the fungus Aspergillus violaceofuscus from the marine sponge Reniochalina sp. resulted in the identification of three new cyclopeptides, including a cyclic tetrapeptide violaceotide A (1), an aspochracin-type cyclic tripeptide sclerotiotide L (2), and a new diketopiperazine dimer (3) (Figure 1). Herein, the isolation, structure elucidation and anti-inflammatory studies of the three new cyclic peptides were described.
Figure 1.
Structures of compounds 1-3.
Materials and methods
General experimental procedures
Optical rotations were determined on a Rudolph research analytical autopol VI polarimeter with a 1 dm length cell at room temperature. UV spectra were performed on a Persee TU-1950 UV-VIS spectrophotometer. The NMR spectra were recorded on a Bruker AMX-600 instrument. HRESIMS data were obtained on a Waters Xevo G2-XS Q-Tof mass spectrometer. Reversed-phase HPLC was performed on Waters X-Bridge C18 (5 μm) columns with a Waters 1525 separation module equipped with a Waters 2998 photodiode array detector. MPLC was accomplished using a Interchim PuriFlash 450 chromatography system. Silica gel 60 (200–300 mesh; Yantai, China), Sephadex LH-20 (18–110 μm, Pharmacia Co.) and ODS (50 μm, YMC Co.) were used for column chromatography.
Fungal strain and fermentation
The fungus Aspergillus violaceofuscus was isolated from the inner part of the marine sponge Reniochalina sp. collected from the Xisha Islands in the South China Sea. The sample was deposited at the Research Center for Marine Drugs, School of Medicine, Shanghai Jiao Tong University. This strain was identified based on the morphology analyze and ITS gene sequencing (GenBank accession No. FJ491681).
The strain was cultivated on potato dextrose agar at 28°C for 7 days. Large scale fermentation was carried out in 50 erlenmeyer flasks (2 L) each containing 80 g of rice and 120 mL of distilled H2O with 0.3% (m/v) peptone. Each flask was inoculated with 20 mL of cultured broth and incubated under static conditions at room temperature for 40 days.
Extraction and isolation
The fermented substrate was exhaustively extracted with ethyl acetate to provide the residue (26.0 g) after removal of the organic solvent under reduced pressure.
The EtOAc extract was fractionated by vacuum liquid chromatography on silica gel (200–300 mesh) using CH2Cl2/MeOH gradient elution (500:1–0:1, v/v) to give eleven fractions A–K. Fraction H (6.1 g) was separated by column chromatography (CC) over Sephadex LH-20 eluted with MeOH to afford subfractions H1–H9. Subfraction H3 (1.5 g) was applied to medium pressure liquid chromatography (MPLC) on ODS, eluted with a gradient of 10 to 100% (v/v) MeCN in H2O, to give H3A–H3F. H3D was subjected to reversed-phase HPLC with an elution of 55% MeOH in H2O to give 1 (2.0 mL/min, tR = 23.0 min, 8.6 mg). H3C (31.3 mg) was purified by semi-preparative reversed-phase HPLC eluted with 18% MeCN in H2O to yield 2 (2.0 mL/min, tR = 37.7 min, 2.0 mg). Subfraction H2 (1.82 g) was applied to MPLC on ODS, eluted with a gradient of 10 to 100% (v/v) MeCN in H2O, to give Fr. H2A–H2H. Fraction H2F (169 mg) was then purified by semi-preparative RP-HPLC eluted with 37% MeCN in H2O resulted in the isolation of 3 (2.0 mL/min, tR = 30.5 min, 1.8 mg).
Violaceomide A (1): white amorphous powder; [α]25 D −230 (c 0.6, MeOH); UV (MeOH) λmax (log ε) 220 (3.26), 276 (1.31) nm; HRESIMS m/z 477.2719 [M + H]+ (calcd for C24H37N4O6, 477.2713); 1H and 13C NMR data, Table 1.
Table 1.
1H (600 MHz) and 13C NMR (150 MHz) Data for 1 in Pyridine-d5.
| Position | δC | δH, mult. (J in Hz) | Position | δC | δH, mult. (J in Hz) |
|---|---|---|---|---|---|
| N-Me-Ala | 12 | 65.3, CH | 4.37, dd (8.9, 2.4) | ||
| 1 | 173.6, C | 13 | 68.1, CH | 4.78, m | |
| 2 | 55.1, CH | 4.74, m | 14 | 22.2, CH3 | 1.40, d (6.3) |
| 3 | 17.0, CH3 | 1.46, d (7.1) | 12-NH | 7.20, brs | |
| 4 | 30.8, CH3 | 3.32, s | O-Me-Tyr | ||
| Ile | 15 | 173.8, C | |||
| 5 | 171.4, C | 16 | 55.9, CH | 4.47, m, overlapped | |
| 6 | 55.7, CH | 5.14, dd (10.0, 7.6) | 17 | 35.7, CH2 | 3.81, m; 3.63, m |
| 7 | 37.6, CH | 2.37, m | 18 | 132.7, C | |
| 8 | 17.6, CH3 | 1.16, d (6.5) | 19/23 | 131.9, CH | 7.31, d (8.0) |
| 9 | 25.2, CH2 | 2.00, m; 1.42, m | 20/22 | 114.9, CH | 7.03, d (8.2) |
| 10 | 12.4, CH3 | 0.94, t (7.4) | 21 | 159.6, C | |
| 6-NH | 8.80, brs | 24 | 55.9, CH3 | 3.78, s | |
| Thr | 16-NH | 9.30, brs | |||
| 11 | 173.2, C |
Sclerotiotide L (2): pale yellow amorphous powder; [α]25 D −92 (c 0.5, MeOH); UV (MeOH) λmax (log ε) 216 (3.84), 258 (3.98) nm; HRESIMS m/z 481.3042 [M + H]+ (calcd for C24H41N4O6, 481.3026); 1H and 13C NMR data, Table 2.
Table 2.
1H (600 MHz) and 13C NMR (150 MHz) Data for 2 in CDCl3.
| Position | δC | δH, mult. (J in Hz) | Position | δC | δH, mult. (J in Hz) |
|---|---|---|---|---|---|
| Ala | 10-NH | 6.53, d (7.2) | |||
| 1 | 171.5, C | 11 | 28.6, CH2 | 2.39, m; 1.60, m | |
| 2 | 55.2, CH | 4.59, q (7.1) | 12 | 21.9, CH2 | 1.66, m; 1.57, m |
| 3 | 17.0, CH3 | 1.51, d (7.1) | 13 | 39.7, CH2 | 3.38, m; 3.06, m |
| N-CH3 | 29.9, CH3 | 3.06, s | 13-NH | 5.68, brs | |
| Val | Fatty acid | ||||
| 4 | 169.2, C | 1′ | 164.8, C | ||
| 5 | 58.8, CH | 5.11, d (10.5) | 2′ | 124.2, CH | 5.92, d (15.0) |
| 6 | 27.0, CH | 2.43, m | 3′ | 140.3, CH | 7.23, dd (15.0, 10.8) |
| 7 | 20.0, CH3 | 0.92, d (6.3) | 4′ | 132.1, CH | 6.36, dd (15.4, 11.0) |
| 8 | 18.0, CH3 | 0.74, d (6.8) | 5′ | 137.7, CH | 6.00, dd (15.4, 7.8) |
| N-CH3 | 30.4, CH3 | 2.95, s | 6′ | 85.5, CH | 3.62, dd (7.8, 3.7) |
| Orn | 7′ | 69.5, CH | 3.90, dd (6.5, 3.7) | ||
| 9 | 173.1, C | 8′ | 18.0, CH3 | 1.12, d (6.5) | |
| 10 | 49.7, CH | 4.98, t (7.2) | 9′ | 57.1, CH3 | 3.32, s |
Compound (3): White powder. [α]25 D +530.0 (c 0.3, MeOH); UV (MeOH) λmax (log ε) 239 (3.58), 300 (1.55) nm; HRESIMS m/z 597.3177 [M + H]+ (calc. for C34H41N6O4 597.3189). 1H and 13C NMR data, Table 3.
Table 3.
1H (600 MHz) and 13C NMR (150 MHz) Data for 3 in CDCl3.
| Position | δC | δH, mult. (J in Hz) | Position | δC | δH, mult. (J in Hz) |
|---|---|---|---|---|---|
| 2/2′ | 80.5, CH | 4.94, s | 12/12′ | 37.2, CH2 | 3.18, dd (14.0, 9.1) |
| 3/3′ | 59.7, C | 13/13′ | 168.5, C | ||
| 4/4′ | 130.0, C | 14/14′ | 5.93, s | ||
| 5/5′ | 124.9, CH | 7.34, d (7.5) | 15/15′ | 56.3, CH | 3.77, m |
| 6/6′ | 119.9, CH | 6.81, t (7.5) | 16/16′ | 168.2, C | |
| 7/7′ | 129.8, CH | 7.15, t (7.6) | 17/17′ | 41.8, CH2 | 1.48, m; 2.82, dd (14.0, 8.6) |
| 8/8′ | 110.3, CH | 6.65, d (7.9) | 18/18′ | 24.5, CH | 1.64, m |
| 9/9′ | 148.8, C | 19/19′ | 23.1, CH3 | 0.89, d (6.5) | |
| 11/11′ | 55.8, CH | 3.99, t (8.8) | 20/20′ | 21.3, CH3 | 0.87, d (6.5) |
Advanced Marfey's analysis of compound 1
Compound 1 (1 mg) were hydrolyzed in HCl (6 M; 1 mL) for 18 h at 110°C. The solutions were then evaporated to dryness and redissolved in H2O (200 μL). The aqueous hydrolysate was added with 1% (w/v) solution of 1-fluoro-2,4-dinitrophenyl-5-L-leucinamide (L-FDLA, 100 μL) in acetone and 1 M NaHCO3 (40 μL). After treating at 45°C for 90 min, the reactions were quenched by the addition of HCl (1 M, 40 μL). Appropriate amino acid standards were treated with L-FDLA and D-FDLA as described above and yielded the L-FDLA and D-FDLA standards. Marfey's derivatives of 1 was subjected to UPLC-MS selected ion chromatography on a reversed-phase column (Waters ACQUITY HS T3 column; 1.8 μm, 2.1 × 100 mm) with a linear gradient from 10 to 60% aqueous CH3CN containing 0.1% formic acids over 18 min and their retention times were compared with those from the authentic standard derivatives.
Advanced Marfey's analysis of compound 2
Compound 2 (1 mg) were hydrolyzed in HCl (6 M; 1 mL) for 20 h at 110°C. The solutions were then evaporated to dryness and redissolved in H2O (200 μL). The aqueous hydrolysate was divided into two equal portions. One portion was treated with 1% (w/v) solution of 1-fluoro-2,4-dinitrophenyl-5-D-leucinamide (D-FDLA, 100 μL) in acetone and 1 M NaHCO3 (40 μL). The second portion was treated with a racemic mixture of a 1% (w/v) solution of 1-fluoro-2,4-dinitrophenyl-5-D-leucinamide (D-FDLA, 50 μL) in acetone, 1% (w/v) solution of 1-fluoro-2,4-dinitrophenyl-5-L-leucinamide (L-FDLA, 50 μL) in acetone, and 1 M NaHCO3 (40 μL). Both mixtures were heated at 45°C for 90 min and the reactions were quenched by the addition of HCl (1 M, 40 μL). The aliquots were subjected to HPLC-MS selected ion chromatography on a reversed-phase column (Waters XBridge C18 column; 5 μm, 4.6 × 250 mm; 1.0 mL/min) with a linear gradient from 10 to 80% aqueous CH3CN containing 0.1% formic acids over 30 min according to the advanced Marfey's method. The retention times and ESIMS product ions (tR in min, m/z [M + H]+) of the D-FDLA mono-derivatized amino acids in the hydrolysate of the first portion were observed to be Orn (13.1, 427.5), N-Me-Ala (20.4, 398.5), and N-Me-Val (24.3, 426.4), while the reaction with racemic D/L-FDLA in the second portion gave rise to two peaks for each corresponding amino acid moiety. The retention times and ESIMS product ions (tR1/tR2, min, m/z [M + H]+) were observed to be Orn (13.1/14.1, 427.5), N-Me-Ala (20.1/20.4, 398.5), and N-Me-Val (22.1/24.3, 426.4). Consequently, the absolute configuration of the amino acids in the hydrolysate of 2 was confirmed as L-Orn, N-Me-L-Ala, and N-Me-L-Val.
Marfey's analysis of compound 3
Compound 3 (0.4 mg) was dissolved in 6 N HCl (1 mL) and heated at 110°C for 24 h. Then, the solvent was evaporated under reduced pressure and resuspended in 50 μL of H2O. The hydrolysates were treated with 200 μL of 1% (w/v) 1-fluoro-2,4-dinitrophenyl-5-L-leucinamide (FDLA) in acetone and 40 μL of 1.0 N NaHCO3. The reaction mixtures were heated at 45°C for 2 h, cooled to room temperature, and then neutralized with 40 μL of 1 N HCl. Standard D-Leu and D/L-Leu were derivatised in a similar fashion separately. The derivatives of the hydrolysates and the standard amino acids were analyzed by LC-MS selected ion chromatography on a reversed-phase column (Waters XBridge C18 column; 5 μm, 4.6 × 250 mm; 1.0 mL/min) with a linear gradient from 10 to 100% aqueous CH3CN containing 0.1% formic acids over 30 min. The retention times for FDLA derivatives of standard D-Leu and L-Leu were 21.8 and 18.7 min, respectively, while this for FDLA derivatives of compound 3 were 21.8 min.
Anti-inflammatory assay
THP-1 (a human acute monocytic leukemia cell line) cells (CCTCC) were maintained in RPMI-1640 supplemented with 10% (v/v) FBS and 0.05 mmol/L 2-mercaptoethanol at 37°C in a 5% CO2 and humidified environment. THP-1 cells (5 × 105/mL) were differentiated using 160 nmol/L PMA for 36 h. Differentiation of PMA-treated cells was enhanced by removing the PMA-containing media and the cells were incubated in FBS free, fresh RPMI 1640 for a further 12 h, and then stimulated with compounds or/and LPS at the indicated concentrations and time periods.
Cytokines IL-6, IL-10, MCP-1, and TNF-α in the culture media of THP-1 cells treated with 10 μM compounds or/and 0.1 μg/mL LPS were determined by flow cytometry using the Human Inflammation Cytometric Bead Array (CBA) according to the instruction of the manufacturer (BD Biosciences, San Jose, CA, USA). Cytokine levels were measured on a FACSCalibur flow cytometer (BD Biosciences Pharmingen). The concentrations were assessed by using FCAP Array software.
Results and discussion
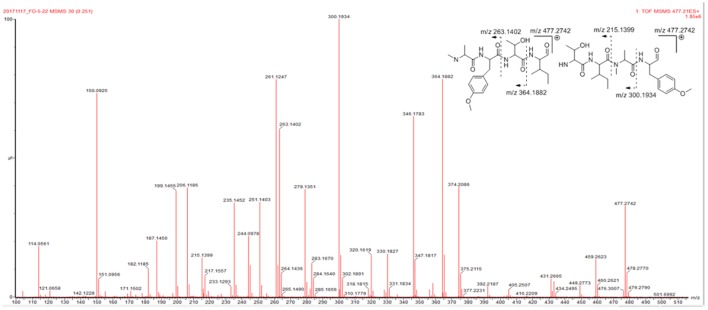
Compound 1 was isolated as a White, amorphous powder, which possessed a molecular formula of C24H36N4O6 deduced from the pseudomolecular ion peak at m/z 477.2719 [M + H]+ in its HRESIMS (Supplementary Figure 19). The signal distribution pattern observed in the 1H and 13C NMR spectrum (pyridine-d5) (Supplementary Figures 1–2), which showed three exchangeable amide NH signals (δH 9.30, 8.80, 7.20), four amide carbonyls (δC 173.8, 173.6, 173.2, and 171.4), four characteristic α-methine signals (δH/C 5.14/55.7, 4.74/55.1, 4.47/55.9, and 4.37/65.3) and one N-methyl (δH/C 3.32/30.8), indicated the characteristic of a peptide. Combined analysis of the 2D NMR spectra (Supplementary Figures 3–6) revealed the structures of four amino acid residues, including alanine (Ala), isoleucine (Ile), threonine (Thr), and one tyrosine (Tyr). An HMBC correlation from 4-N-CH3 (δH 3.32) to Ala C-2 (δC 55.1) indicated that the Ala residue was N-methylated. The 24-OCH3 linked to the benzene ring at C-21 was supported by the HMBC correlation from H3-24 (δH 3.78) to C-21 (δC 159.6). The assignment of the amino acid sequence was carried out by a combination of HMBC, NOESY (Figure 2), and MS/MS analysis. The HMBC correlations from NMeAla H3-4 (δH 3.32) to Ile C-5 (δC 171.4) and from Ile H-6 (δH 5.14) to Thr C-11 (δC 173.2), suggesting a partial sequence of NMeAla-Ile-Thr. The NOESY correlation between OMeTyr NH (δH 9.30) and NMeAla H3-3 (δH 1.46) extended this sequence to OMeTyr-NMeAla-Ile-Thr. The 9° of unsaturation and the molecular formula suggested that 1 was a cyclic peptide. Therefore, the cyclic tetrapeptide ring was closed between OMeTyr and Thr. In addition, the amino acid sequence of 1 was confirmed by mass fragmentation analysis using a quadrupole-time-of-flight (Q-TOF) tandem mass spectrometer (Figure 3). Consequently, the planar structure of 1 was elucidated as a cyclic tetrapeptide with the sequence cyclo-(Thr-O-MeTyr-N-MeAla-Ile).
Figure 2.
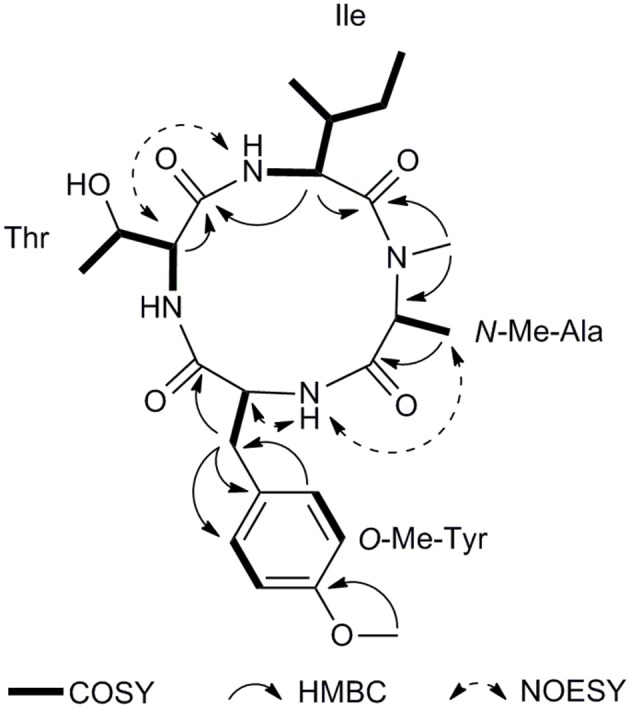
Key COSY, HMBC, and NOESY correlations of 1.
Figure 3.
MS/MS spectrum of 1.
The absolute configurations of the amino acids were determined by the advanced Marfey's method after acid hydrolysis (Fujii et al., 1997). Compound 1 was hydrolyzed and then derivatized with L-FDLA. The UPLC-MS comparison between Marfey's derivatives of the hydrolysate of 1 and appropriate amino acid standards assigned the L configurations for Thr, Tyr, N-Me-Ala, and Ile (Supplementary Figure 23). The final structure of 1 was elucidated as cyclo-[L-Thr–L-O-Me-Tyr–L-N-Me-Ala–L-Ile] and named as violaceotide A.
Compound 2 was isolated as a pale yellow amorphous powder. The molecular formula can be determined as C24H40N4O6 by HRESIMS ion peak at m/z 481.3042 [M + H]+ (calcd for C24H41N4O6, 481.3026) (Supplementary Figure 20). The 1H NMR spectrum of 2 showed an amide NH proton (δH 6.53), two N-methyl protons (δH 2.95 and 3.06), and three characteristic α-methine signals (δH 4.59, 4.98, and 5.11), indicating a tripeptide structure. The 13C NMR spectrum exhibited a total of 24 carbon resonances, including four amide carbonyl carbons, ten methine carbons, three methene carbons, and seven methyl carbons. The obvious difference in the NMR spectra between compound 2 and sclerotiotide H (Zheng et al., 2010) was the appearance of an additional O-methyl group at δH 3.32 (H-9′) and δC 57.1 (C-9′). The HMBC correlations (Figure 4) from H3-9′ to C-6′ (δC 85.5) revealed that the O-methyl was linked to fatty acid chain at position 6′ (Supplementary Figures 7–12). The geometries of the Δ2′, 3′ and Δ4′, 5′ olefins were identified as 2′E and 4′E, by the proton spin coupling constants of (15.0 Hz) and (15.4 Hz). In order to determine the absolute configurations of the amino acid residues of 2, advanced Marfey's method was utilized (Fujii et al., 1997). HPLC-MS analysis of derivatives of the hydrolysates with D-FDLA and D/L-FDLA indicated that the amino acids were NMe-L-Ala, NMe-L-Val, and L-Orn (Supplementary Figure 24). Thus, compound 2 was elucidated as (2′E,4′E)-cyclo-[(NMe-L-Ala)-(NMe-L-Val)-(Nα-6′-methoxy-7′-hydroxyocta-2′,4′-dienoyl-L-Orn)] and named as sclerotiotide L.
Figure 4.
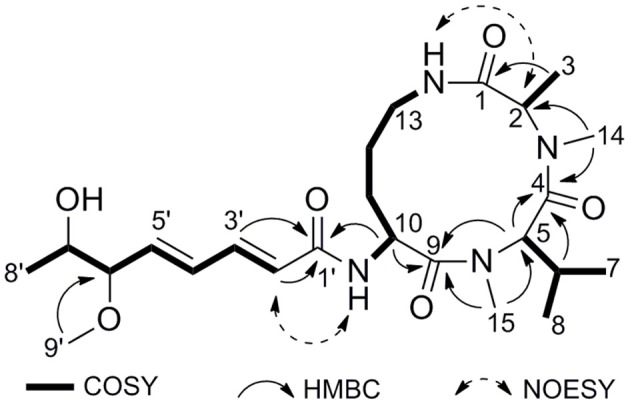
Key COSY, HMBC, and NOESY correlations of 2.
Compound 3 was isolated as a white amorphous powder. Its molecular formula was determined as C34H40N6O4 on the basis of the pseudomolecular ion peak at m/z 597.3177 [M + H]+ (calcd for C34H41N6O4, 597.3189) in the HRESIMS (Supplementary Figure 21), requiring 18 degrees of unsaturation. The NMR spectra of 3 revealed 17 carbon signals, indicating that 3 would be a symmetric homodimer which was further confirmed by its half MS fragment ion peak at m/z 298.2 (Supplementary Figure 22).
In the 1H NMR spectrum, four aromatic signals at δH 7.34 (d, J = 7.5 Hz, H-5/H-5′), 7.15 (t, J = 7.6 Hz, H-7/H-7′), 6.81 (t, J = 7.5 Hz, H-6/H-6′), and 6.65 (d, J = 7.9 Hz, H-8/H-8′) and a proton at 4.94 (s, 1H, H-2/H-2′) were observed, which suggested a indoline moiety.16 In addition, two methyl signals were also observed at δH 0.89 (d, J = 6.5 Hz, H3-19/H3-19′) and 0.87 (d, J = 6.5 Hz, H3-20/H3-20′). The 13C NMR spectrum exhibited a total of 17 carbon resonances, including five quaternary carbons, eight methine carbons, two methene carbons, and two methyl carbons. Two 13C NMR resonances at δ 168.5 (C-13/C-13′) and δ 168.2 (C-16/C-16′) were characteristic signals of lactam carbonyls. The HMBC correlations from H-17 (δH 1.48) to C-15 (δC 56.3), and C-16 (δC 168.2) and COSY correlations of H-14 (δH 5.93)/H-15 (δH 3.77)/H-17/H-18 (δH 1.64)/H-19 and H-18/H-20 established a leucine unit (Figure 5). A tryptophan moiety can be concluded from the 1H NMR signals and the key HMBC correlations from H-12 (δH 3.18) to C-2 (δC 80.5), C-3 (δC 59.7), C-11 (δC 55.8), and C-13, from H-2 to C-3, and C-9 (δC 148.8). Ultimately, extensive analysis of the 2D NMR data and comparison of the spectroscopic data with the reported literature (Ovenden et al., 2004) allowed identifying the planar structure of 3 as shown (Supplementary Figures 13–18).
Figure 5.
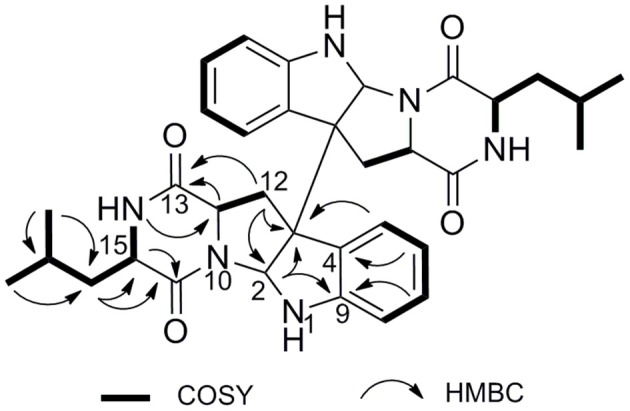
Key HMBC and COSY correlations of 3.
The relative configurations of 3 were deduced from the observed NOESY correlations (Figure 6). The key NOESY correlations of H-11/H-17 and H-11/ H-2, indicated that these protons were on the same face. The NOESY correlations of H-5′ with H-11 and H-11 with H-2 suggested cis-fused ring junction at C-2 and C-3. Marfey's method (Cho et al., 2018) was employed to determine the absolute configuration at C-15. Compound 3 was hydrolyzed and derivatized with L-FDLA and analyzed by LC-MS to the establishment of absolute configuration of Leu residues. By comparing the retention times of authentic standards of L- and D- forms of Leu, the hydrolysate was identified to contain a unit of D-Leu (Supplementary Figure 25). Therefore, the absolute configurations of 3 were assigned as 2R, 3R, 11S, 15R (Figure 1).
Figure 6.
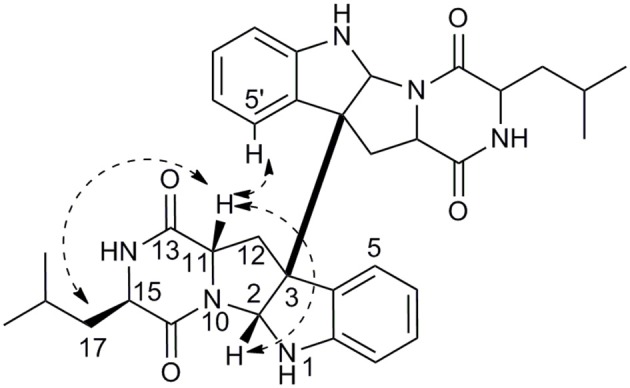
Key NOESY correlations of 3.
To the best of our knowledge, compound 3 has the same planar structure as a unnamed and ambiguous compound without any spectroscopic data or optical rotation published on a patent (Masashi et al., 1995). Our compound possesses the different stereochemistry with it. Therefore, 3 was reported as a new diketopiperazine dimer herein.
Compounds 1–3 were evaluated their inhibitory activities against the production of four cytokines levels in the serum of human acute monocytic leukemia cell line THP-1 by using the human inflammation cytometric bead array (CBA) assay (Table 4). The cytokines, including IL-6, IL-10, MCP-1, TNF-α in this study, are pivotal mediators that contribute to inflammation and various related diseases (Liu et al., 2017; Wu et al., 2017). Notably, treatment of THP-1 cells by LPS showed a significant elevation in the secretion of the cytokines (P < 0.01). Results showed that the THP-1 cells pretreated with compounds 1 and 3 showed a significant decrease in the LPS-induced expression of IL-10 with inhibitory rates of 84.3 and 78.1% (P < 0.01), respectively. These compounds did not show cytotoxicity against THP-1 cells after 24 h treatment.
Table 4.
The inhibitory rates of compounds 1-3 against the cytokines expression of LPS-induced THP-1 cells at concentration of 10 μM.
| Compound | IL-6 (%) | IL-10 (%) | MCP-1 (%) | TNF-α (%) |
|---|---|---|---|---|
| 1 | 45.9 | 84.3 | 32.9 | 64.2 |
| 2 | 28.0 | 23.6 | 40.5 | 61.5 |
| 3 | 51.2 | 78.1 | 40.0 | 63.1 |
Conclusions
From the marine sponge-derived fungus Aspergillus violaceofuscus, three new cyclic peptides were obtained. Aspochracin-type cyclic tripeptide sclerotiotide L (2) and a diketopiperazine dimer (3) showed anti-inflammatory activity against IL-10 expression of the LPS-induced THP-1 cells, which indicated that the marine sponge-derived microorganism are a fertile source of compounds with novel structures and significant bioactivities.
Author contributions
JL, BG, and LY: performed the experiments; JL: identified the structures and analyzed the data; HL and FY: conceived and designed the experiments; JL and FY: wrote the paper. All authors listed have approved the work for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was supported by the National Natural Science Fund for Distinguished Young Scholars of China (81225023), the National Natural Science Fund of China (Nos. U1605221, 81741151, and 41476121), Shanghai Municipal Commission of Health and Family Planning (20140388), and the Fund of the Science and Technology Commission of Shanghai Municipality (15431900900).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2018.00226/full#supplementary-material
References
- Amagata T., Morinaka B. I., Amagata A., Tenney K., Valeriote F. A., Lobkovsky E., et al. (2006). A chemical study of cyclic depsipeptides produced by a sponge-derived fungus. J. Nat. Prod. 69, 1560–1565. 10.1021/np060178k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J., Zhang X.-Y., Xu X.-Y., He F., Nong X.-H., Qi S.-H. (2013). New cyclic tetrapeptides and asteltoxins from gorgonian-derived fungus Aspergillus sp. SCSGAF 0076. Tetrahedron 69, 2113–2117. 10.1016/j.tet.2013.01.021 [DOI] [Google Scholar]
- Cho K. H., Sohn J. H., Oh H. (2018). Isolation and structure determination of a new diketopiperazine dimer from marine-derived fungus Aspergillus sp. SF-5280. Nat. Prod. Res. 32, 214–221. 10.1080/14786419.2017.1346642 [DOI] [PubMed] [Google Scholar]
- Daletos G., Kalscheuer R., Koliwer-Brandl H., Hartmann R., de Voogd N. J., Wray V., et al. (2015). Callyaerins from the Marine Sponge Callyspongia aerizusa: cyclic peptides with antitubercular activity. J. Nat. Prod. 78, 1910–1925. 10.1021/acs.jnatprod.5b00266 [DOI] [PubMed] [Google Scholar]
- Ding L. J., Yuan W., Liao X. J., Han B. N., Wang S. P., Li Z. Y., et al. (2016). Oryzamides A-E, Cyclodepsipeptides from the Sponge-Derived Fungus Nigrospora oryzae PF18. J. Nat. Prod. 79, 2045–2052. 10.1021/acs.jnatprod.6b0034 [DOI] [PubMed] [Google Scholar]
- Fujii K., Ikai Y., Oka H., Suzuki M., Harada K.-I. (1997). A nonempirical method using LC/MS for determination of the absolute configuration of constituent Amino Acids in a Peptide: combination of Marfey's method with mass spectrometry and its practical application. Anal. Chem. 69, 5146–5151. [Google Scholar]
- Fukuhara K., Takada K., Okada S., Matsunaga S. (2015). Nazumazoles A-C, cyclic pentapeptides dimerized through a disulfide bond from the marine sponge Theonella swinhoei. Org. Lett. 17, 2646–2648. 10.1021/acs.orglett.5b01020 [DOI] [PubMed] [Google Scholar]
- Gu W. X., Cueto M., Jensen P. R., Fenical W., Silverman R. B. (2007). Microsporins A and B: new histone deacetylase inhibitors from the marine-derived fungus Microsporum cf. gypseum and the solid-phase synthesis of microsporin A. Tetrahedron 63, 6535–6541. 10.1016/j.tet.2007.04.025 [DOI] [Google Scholar]
- Ireland C. M., Jr., Durso A. R., Newman R. A., Hacker M. P. (1982). Antineoplastic cyclic peptides from the marine tunicate Lissoclinum patella. J. Org. Chem. 47, 1807–1811. [Google Scholar]
- Jang J. P., Nogawa T., Futamura Y., Shimizu T., Hashizume D., Takahashi S., et al. (2017). Octaminomycins A and B, Cyclic Octadepsipeptides Active against Plasmodium falciparum. J. Nat. Prod. 80, 134–140. 10.1021/acs.jnatprod.6b00758 [DOI] [PubMed] [Google Scholar]
- Liu J. T., Wu W., Cao M. J., Yang F., Lin H. W. (2017). Trienic alpha-pyrone and ochratoxin derivatives from a sponge-derived fungus Aspergillus ochraceopetaliformis. Nat. Prod. Res.. 10.1080/14786419.2017.1402325 [DOI] [PubMed] [Google Scholar]
- Luo S., Krunic A., Kang H. S., Chen W. L., Woodard J. L., Fuchs J. R., et al. (2014). Trichormamides A and B with antiproliferative activity from the cultured freshwater Cyanobacterium Trichormus sp. UIC 10339. J. Nat. Prod. 77, 1871–1880. 10.1021/np5003548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masashi H., Hiroshi M., Mitsuyoshi S., Toshie H., Hidenor Y. (1995). Cholecystokinin B/Gastrin Receptor Antagonist. Japan patent application JP 1994–119123.
- Ovenden S. P., Sberna G., Tait R. M., Wildman H. G., Patel R., Li B., et al. (2004). A diketopiperazine dimer from a marine-derived isolate of Aspergillus niger. J. Nat. Prod. 67, 2093–2095. 10.1021/np0497494 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., Bruijn I. d., Nybroe O., Ongena M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1061. 10.1111/j.1574-6976.2010.00221.x [DOI] [PubMed] [Google Scholar]
- Randazzo A., Bifulco G., Giannini C., Bucci M., Debitus C., Cirino G., et al. (2001). Halipeptins A and B: two novel potent anti-inflammatory cyclic depsipeptides from the Vanuatu marine sponge Haliclona species. J. Am. Chem. Soc. 123, 10870–10876. 10.1021/ja010015c [DOI] [PubMed] [Google Scholar]
- Sieber S. A., Marahiel M. A. (2005). Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105, 715–738. 10.1021/cr0301191 [DOI] [PubMed] [Google Scholar]
- Teta R., Marteinsson V. T., Longeon A., Klonowski A. M., Groben R., Bourguet-Kondracki M. L., et al. (2017). Thermoactinoamide A, an Antibiotic Lipophilic Cyclopeptide from the Icelandic Thermophilic Bacterium Thermoactinomyces vulgaris. J. Nat. Prod. 80, 2530–2535. 10.1021/acs.jnatprod.7b00560 [DOI] [PubMed] [Google Scholar]
- Wu W., Zhen Z., Niu T., Zhu X., Gao Y., Yan J., et al. (2017). kappa-Carrageenan Enhances Lipopolysaccharide-Induced Interleukin-8 Secretion by Stimulating the Bcl10-NF-kappaB Pathway in HT-29 Cells and Aggravates C. freundii-Induced Inflammation in Mice. Med. Inflamm. 2017:8634865. 10.1155/2017/8634865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. J., Liao X. J., Xu S. H., Diao J. Z., Du B., Zhou X. L., et al. (2008). Isolation, Structure Determination, and Synthesis of Galaxamide, A Rare Cytotoxic Cyclic Pentapeptide from a Marine Algae Galaxaura filamentosa. Org. Lett. 10, 4569–4572. 10.1021/ol801799d [DOI] [PubMed] [Google Scholar]
- Yu Z., Lang G., Kajahn I., Schmaljohann R., Imhoff J. F. (2008). Scopularides A and B, cyclodepsipeptides from a marine sponge-derived fungus, Scopulariopsis brevicaulis. J. Nat. Prod. 71, 1052–1054. 10.1021/np070580e [DOI] [PubMed] [Google Scholar]
- Zhang H. J., Yi Y. H., Yang G. J., Hu M. Y., Cao G. D., Yang F., et al. (2010). Proline-containing cyclopeptides from the marine sponge Phakellia fusca. J. Nat. Prod. 73, 650–655. 10.1021/np9008267 [DOI] [PubMed] [Google Scholar]
- Zheng J., Xu Z., Wang Y., Hong K., Liu P., Zhu W. (2010). Cyclic tripeptides from the halotolerant fungus Aspergillus sclerotiorum PT06-1. J. Nat. Prod. 73, 1133–1137. 10.1021/np100198h [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.