FIG 7 .
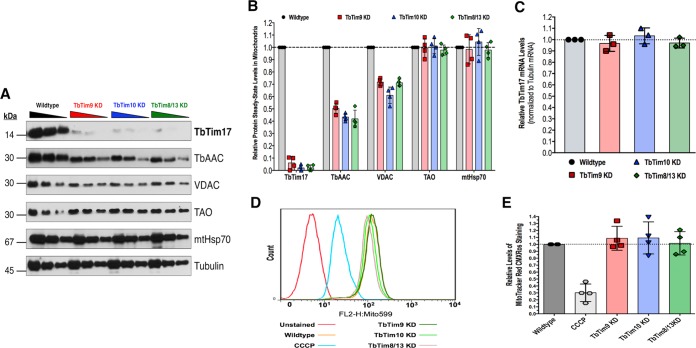
Mitochondrial protein profile of small TbTim knockdown (KD) mitochondria. (A) Western blot analysis of mitochondria (50 µg, 25 µg, and 12.5 µg in decreasing order as indicated by the triangular symbol) isolated from wild-type, TbTim9 RNAi (TbTim9 KD), TbTim10 RNAi (TbTim10 KD), and TbTim8/13 RNAi (TbTim8/13 KD) T. brucei cells grown in the presence of doxycycline (1.0 µg/ml) for 96 h using antibodies as indicated. (B) Scatterplot representation of densitometric analysis of the immunoblots using ImageJ software. The intensities of the indicated protein signals for wild-type mitochondria were normalized to the tubulin signal (loading control). The normalized values for the wild-type sample were set to 100% (1.0). Error bars indicate standard deviations of results from four independent experiments. (C) Scatterplot representation of qRT-PCR analysis of the levels of TbTim17 transcript in the wild-type and small TbTim RNAi cells. Tubulin was used as the internal control. The relative levels of TbTim17 mRNA in small TbTim RNAi cells were calculated by considering the levels in wild-type parasites to represent 1.0. Values were calculated from three independent replicates. Error bars indicate standard deviations. (D) Effect of small TbTim knockdown on MitoTracker Red staining. Cells were treated with doxycycline for 96 h, stained with MitoTracker Red, and fixed with 0.37% paraformaldehyde, and fluorescence intensity was measured at 599 nm by fluorescence-activated cell sorter (FACS) analysis. Wild-type cells pretreated with 50 µM CCCP for 15 min before MitoTracker Red staining served as a positive control for depleted mitochondrial membrane potential. (E) Scatterplot representation of quantitation of relative fluorescence intensities indicative of relative strengths of mitochondrial membrane potential. Error bars indicate standard deviations of results from four independent experiments.