Fig. 3.
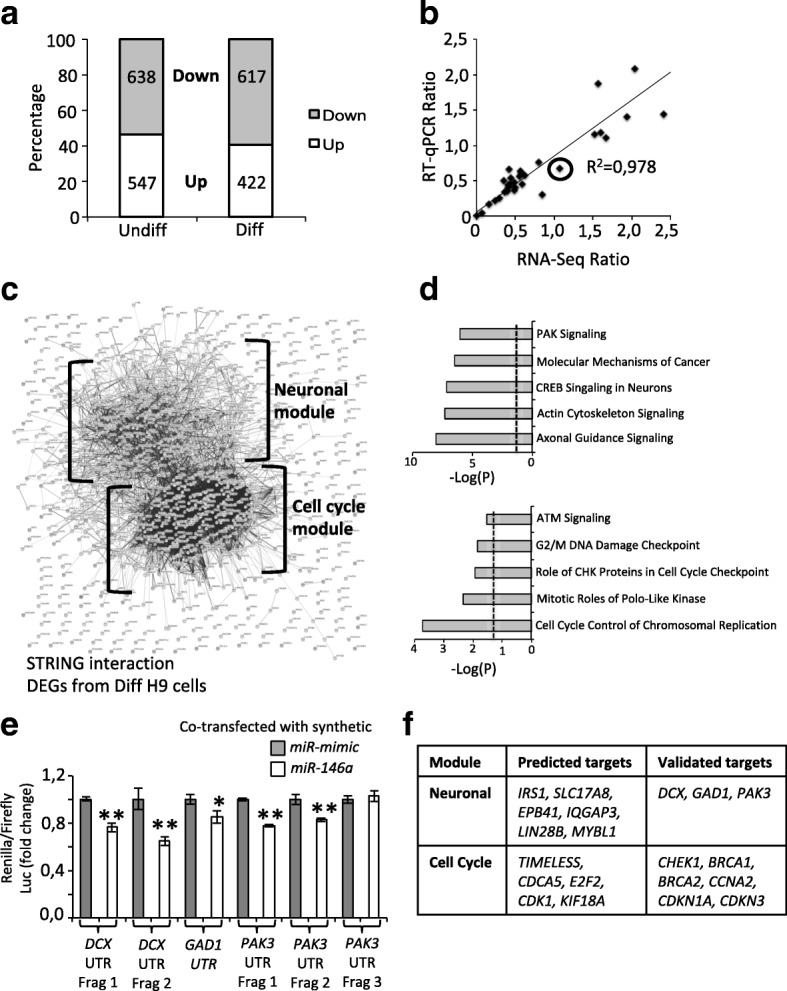
miR-146a targets key genes to promote hNSC differentiation. a Distribution of DEGs identified by RNA-Seq in undifferentiated (Undiff) and differentiated (Diff) conditions. b Graph shows expression ratios of DEGs measured by RNA-Seq and RT-qPCR. Circle indicates only incompatible/non-validated DEGs. c Protein interaction network of all DEGs identified in differentiated cells mapped by STRING. Two modules are identified: Neuronal Module (top) contains 350 genes and Cell Cycle Module (bottom) contains 155 genes. d Top 5 pathways identified by Ingenuity Pathway Analysis of the 2 modules (see Additional file 1: Tables S8, S9 for extended lists). e miR-146a directly targets DCX, GAD1, and PAK3 3′UTR. Due to their size, the 3′UTR of DCX and PAK3 were cloned into 2 and 3 fragments (frag), respectively. Each 3′UTR construct contains at least 3 predicted miR-146a binding sites. Ratio of Renilla/Firefly luciferase (±SD of 3 technical triplicates) was measured in cell lines co-transfected with either the synthetic miR-mimic (control) or the miR-146a. Graphs are representative of 3 independent assays showing the same results. *P < 0.05, **P < 0.01 by Student’s 2 tailed paired t test. f List of known and predicted miR-146a targets that are downregulated in each respective module
