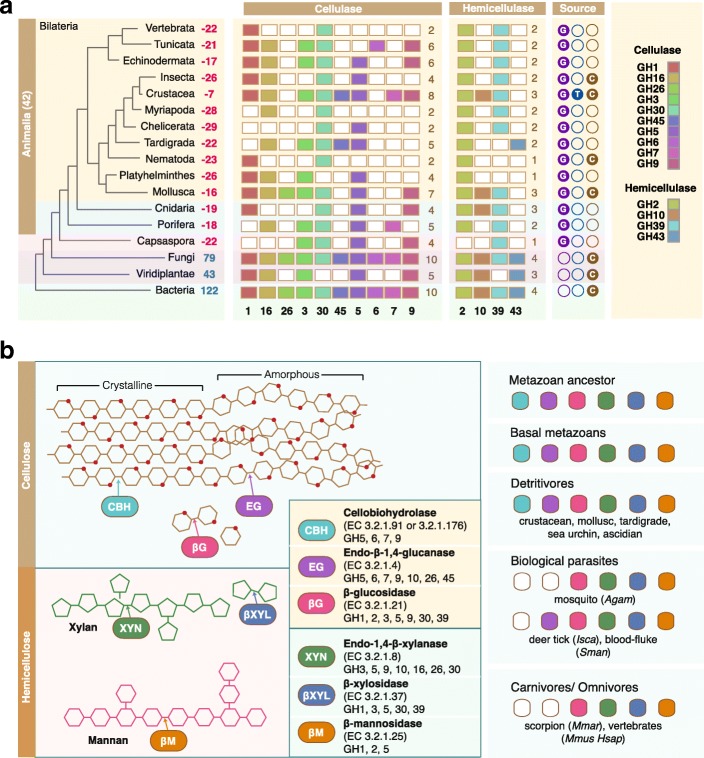
Fig. 4.
Evolution of lignocellulose decomposition machineries in animals. a Comparison of cellulases across the Tree of Life with focus on animals. The number of GH families lost from each taxon are denoted in red beside taxon labels. The number of GH families present in non-animal taxa are denoted in blue. Color boxes indicate the presence of certain cellulase family members while empty boxes indicate the loss of particular members. Data sources used for interpretation are depicted as colored circles; G = genome, T = transcriptome, C = CAZy. b The presence of lignocellulolytic enzymes is related to animal life history and dietary habits. Endoglucanases hydrolyse internal cellulose bonds while cellobiohydrolases cleave cellobiose units at chain ends. Glucosidases hydrolyse cellobiose units to release glucose. Xylan and mannan hemicellulose require xylosidases and mannosidases for their degradation. Animals that adopt detritivorous diets rich in plant biomass possess complete enzymatic suites for efficient lignocellulose decomposition and this is not seen in animals that adopt more specialized diets. Species names are abbreviated with full species names available in Additional file 8: Table S1