Fig. 1.
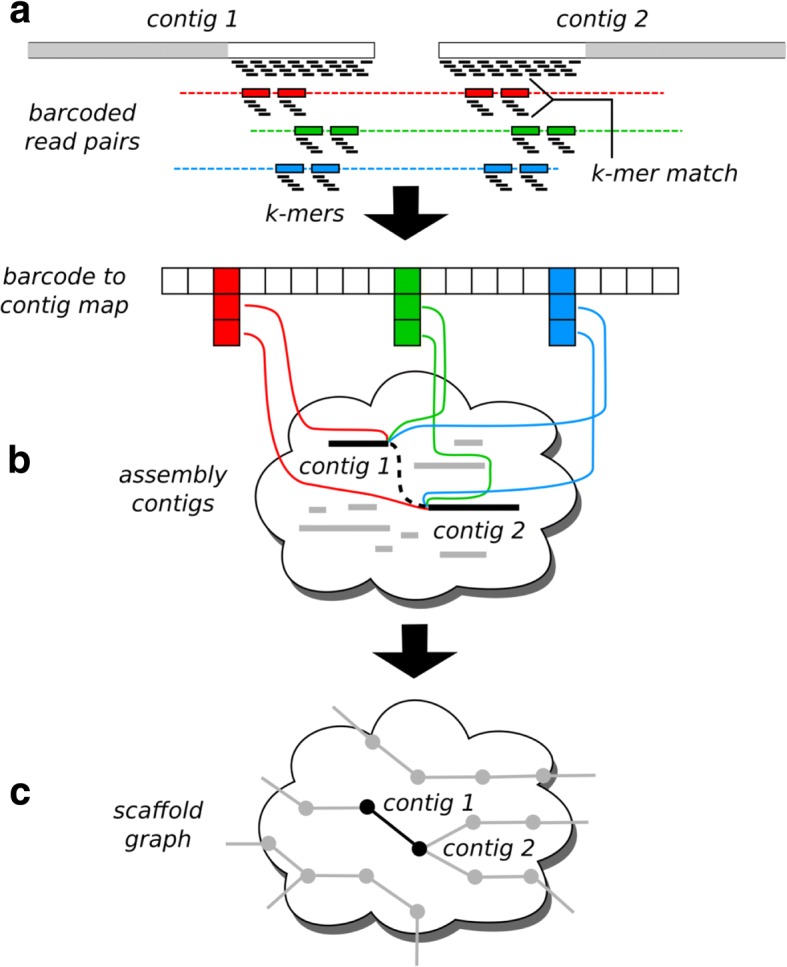
ARKS algorithm. a In the first step, the barcoded Chromium reads are mapped to the ends of the draft assembly sequences (indicated as contigs in the figure) using a kmer-based approach. Reads from three distinct Chromium barcodes are depicted in red, green, and blue, with connecting dashed lines indicating the underlying long DNA molecules for each barcode. The gray regions of the target contigs indicate interior sequence that is masked during mapping. The barcode/contig associations derived from the read mappings are stored in a hash table that maps barcodes to contig ends. b In the second step, we iterate over the barcode-to-contig map and tally the number of barcodes that are shared by each candidate pair of contig ends. c In the third and final step, we generate the output scaffold graph by creating an edge for each candidate pair of contig ends that has greater than a threshold number of shared barcodes (0 by default)
