Fig. 2.
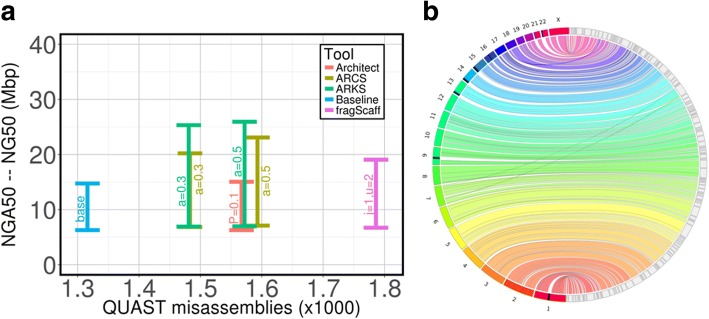
Scaffolding a 10xG Supernova human genome assembly with ARKS. a Comparing the contiguity and accuracy of assemblies scaffolded by ARKS, ARCS, fragScaff and Architect as measured by QUAST. The baseline NA12878 Supernova assembly was scaffolded using ARCS (−c5 -s98 -m50–6000 -z3000 -e30000), ARKS (−c5 -k100 -t8 -j0.5 -m50–6000 -z3000 -e30000), fragScaff (−E30000 -j1 -u2) and Architect (--rc-abs-thr5 --rc-rel-edge-thr0.2, --rc-rel-prun-thr abbreviated to ‘P’). The Y-axes show the range of NGA50 to NG50 lengths to indicate the uncertainty caused by real genomic variations between individual NA24143 and the reference genome GRCh38. b A Circos [24] assembly consistency plot of ARKS (−k100 -j0.5 -c5 -e30000 -z3000 -m50–6000 -r0.05 -a0.5) scaffolding of the baseline NA12878 Supernova assembly. Scaftigs from the largest 123 scaffolds, consisting of 85% (NG85) of the genome, are aligned to GRCh38 with BWA mem [18]. GRCh38 chromosomes are displayed incrementally from 1 (bottom, red) to X (top, fuchsia) on the left while scaffolds (grey with black outlines) are displayed on the right side of the rim. Connections show aligned regions, 1 Mbp and larger, between the genome and scaffolds. Large-scale misassemblies are visible as cross-over ribbons. The black regions on chromosomes indicate reconstruction gaps in the reference. The majority of each chromosome is represented in the final ARKS assembly by no more than 13 assembly scaffolds