Summary
Lactic acid bacteria are widely used for the fermentation of dairy products. While bacterial acidification rates, proteolytic activity and the production of exopolysaccharides are known to influence textural properties of fermented milk products, little is known about the role of the microbial surface on microbe–matrix interactions in dairy products. To investigate how alterations of the bacterial cell surface affect fermented milk properties, 25 isogenic Lactococcus lactis strains that differed with respect to surface charge, hydrophobicity, cell chaining, cell‐clumping, attachment to milk proteins, pili expression and EPS production were used to produce fermented milk. We show that overexpression of pili increases surface hydrophobicity of various strains from 3–19% to 94–99%. A profound effect of different cell surface properties was an altered spatial distribution of the cells in the fermented product. Aggregated cells tightly fill the cavities of the protein matrix, while chaining cells seem to be localized randomly. A positive correlation was found between pili overexpression and viscosity and gel hardness of fermented milk. Gel hardness also positively correlated with clumping of cells in the fermented milk. Viscosity of fermented milk was also higher when it was produced with cells with a chaining phenotype or with cells that overexpress exopolysaccharides. Our results show that alteration of cell surface morphology affects textural parameters of fermented milk and cell localization in the product. This is indicative of a cell surface‐dependent potential of bacterial cells as structure elements in fermented foods.
Introduction
Lactic acid bacteria (LAB) are Gram‐positive bacteria that are generally regarded as safe (GRAS) and are used extensively in food and feed fermentations. They are also found on mucosal surfaces of humans and animals (Stiles and Holzapfel, 1997; Leroy and De Vuyst, 2004). One of the dominant attributes of LAB is the fact that they produce lactic acid as the main metabolic end‐product of fermentation, which leads to acidification and preservation of the fermented product. An additional functionality of many strains is their ability to produce volatile metabolites that are important flavour compounds (Smit et al., 2004, 2005a,b). LAB can also play a significant role in altering textural properties of the end‐products through acidification, proteolytic activity or the production of extracellular polysaccharides (EPS; Smid and Kleerebezem, 2014). One economically very important species of LAB, Lactococcus lactis, is used worldwide in the dairy industry for the production of quark, buttermilk and a huge variety of cheeses.
In general, the matrix of fermented dairy products like yoghurt and cheese consists of aggregated caseins, whey proteins and fat droplets, interspersed with serum and/or whey pockets. In addition, it contains minerals, salts and microorganisms. Interactions between milk components in the matrix and their effect on functionality have been studied extensively (Morell et al., 2014; Dickinson, 2015; Hadde et al., 2015; Joyner and Damiano, 2015; Li and Nie, 2015). For example, apart from protein–protein interactions, the rheological properties of a milk gel also depend on the size and number of fat droplets and their surface composition (Mao and McClements, 2013). When milk is homogenized, the fat droplets are covered and stabilized by milk proteins. Because of the cross‐linking interactions between proteins covering the fat droplets, the milk gel is strong and has a low flow and enhanced stability against the expulsion of serum from the contracting protein matrix, syneresis. Interactions between constituents in the food matrix can be hydrophobic (Jarunglumlert et al., 2015), electrostatic (Cheng et al., 2015), based on hydrogen bonding (Wang et al., 2015), Van der Waals, depletion interaction (Tuinier et al., 2015), the consequence of steric repulsion (Evans et al., 2013) and/or caused by salt bridges (Bosshard et al., 2004).
Relative to the large body of knowledge on interactions between the various milk components (Jelen, 2005; Lucey et al., 2017), little is known about microbe–matrix interactions and the possible interactions between LAB and matrix components of fermented products (Busscher and Weerkamp, 1987; Ploux et al., 2010; Burgain et al., 2014a, 2015). Interactions between microorganisms and milk components could occur via surface properties of both particles (Ly‐Chatain et al., 2010; Burgain et al., 2014b), while different types of interactions can occur at the same time, such as Van der Waals interactions, electrostatic repulsions and attractions, hydrogen bonds, hydrophobic interactions, salt bridges and steric interactions (Ubbink and Schär‐Zammaretti, 2005; Jacobs et al., 2007; Burgain et al., 2013). The surface properties of bacteria are determined by the molecular composition of their cell walls, which can be decorated with (lipo‐)teichoic acids, proteins (Habimana et al., 2007), pili or capsular polysaccharides (CPS; Delcour et al., 1999; Giaouris et al., 2009; Meyrand et al., 2013).
The biodiversity of L. lactis surface properties is very high (Tarazanova et al., 2017). Pili can significantly change the surface of L. lactis upon their overexpression (Tarazanova et al., 2016). Large surface changes can also be introduced by plasmid transfer through conjugation (Neve et al., 1987; Broadbent and Kondo, 1991; Gasson et al., 1995), or by altering the expression of genes such as those encoding autolysins (Buist et al., 1997; Visweswaran et al., 2013) or enzymes involved in galactose utilization (Grossiord et al., 2003). The molecular composition of the cell wall has a big impact on the roughness of the bacterial surface, on bacterial chaining and on cell aggregation (Tarazanova et al., 2016). These properties govern the interactions between LAB and the matrix components (Ly‐Chatain et al., 2010; Burgain et al., 2014a). The production of EPS by a starter culture is known to affect textural properties through the binding of water and thereby increasing milk ropiness. This leads to an increase in milk viscosity and a reduction of syneresis (Hassan et al., 2003; Amatayakul et al., 2006; Prasanna et al., 2013). The charge, stiffness and linearity of EPS molecules impacts on rheological and physical properties of the fermented milk matrix. EPS modification by partial removal of side groups leads to a reduction of efficiency as a thickener (Tuinier et al., 2001).
Contrary to the role of EPS on textural properties of fermented milk, little is known about the influence of bacterial surface properties on interactions with the matrix and their functional consequences on flavour and texture (Ly‐Chatain et al., 2010; Jeanson et al., 2015; Le Boucher et al., 2016). More specific interactions between L. lactis flavours, milk proteins and emulsions were studied by Ly et al., 2008a,b. Here, we studied how bacterial surface properties impact textural parameters of fermented milk. For this, we used 25 isogenic L. lactis strains which differed in known surface properties and cell morphologies. We found that particular surface alterations lead to distinct differences in gel hardness and viscosity of fermented milk. Based on our results, we propose that bacteria can be used as structure elements in fermented foods.
Results
Surface‐altered Lactococcus lactis
To investigate if changes of bacterial cell surface properties affect the functionality of lactococci in fermented products, we performed assays to assess starter culture functionality in 25 isogenic L. lactis strains (Table 1) that differed in surface charge, hydrophobicity, chaining, clumping, attachment to proteins, pili expression and EPS production (Table 2).
Table 1.
Strains and plasmids used in this study
| No | Lactococcus lactis strains | Characteristic | Reference |
|---|---|---|---|
| 1 | NCDO712 | L. lactis dairy isolate, lac +, contains six plasmids – pLP712, pSH71, pSH72, pSH73, pSH74, pNZ712. | Gasson (1983) |
| 2 | NCDO712(pIL253pil) | EryR; harbouring pIL253 with pilin operon spaCB‐spaA‐srtC1‐srtC2 from NCDO712 | Tarazanova et al. (2016) |
| 3 | MG1363 | Plasmid‐cured derivative of L. lactis NCDO712 | Gasson (1983) |
| 4 | MG1363(pIL253pil) | EryR; L.lactis MG1363 harbouring pIL253 with pilin operon spaCB‐spaA‐srtC1‐srtC2 from NCDO712 | Tarazanova et al. (2016) |
| 5 | MG1363(pIL253pil∆1) | EryR; L.lactis MG1363 harbouring pIL253 and pilin operon spaCB‐spaA‐srtC1‐srtC2 from NCDO712 with 1,5 kb internal deletion in spaCB | Tarazanova et al. (2016) |
| 6 | MG1299(pIL253pil) | EryR; L. lactis lac + derivative of NCDO712 which additionally to pLP712 harbouring the pilin operon (spaCB‐spaA‐srtC1‐srtC2) from the same NCDO712 strain | Tarazanova et al. (2016) |
| 7 | MG1299 | Derivative of L. lactis NCDO712, harbours pLP712; lac + | Gasson (1983) |
| 8 | MG1362 | Derivative of L. lactis NCDO712 (described to harbour pSH72) | Gasson (1983) |
| 9 | MG1063 | Derivative of L. lactis NCDO712 (described to harbour pSH73 and pSH72) | Gasson (1983) |
| 10 | MG1261 | Derivative of L. lactis NCDO712 (described to harbour pSH73) | Gasson (1983) |
| 11 | MG1365 | Derivative of L. lactis NCDO712 (described to harbour pSH71) | Gasson (1983) |
| 12 | MG1614 | StrR and RifR; derivative of L. lactis MG1363 | Gasson (1983) |
| 13 | MG1614_clu+ | Lac +. Transconjugant of MG1614, harbours pLP712 from NCDO712 and shows clumping phenotype. | Tarazanova M., Beerthuyzen M., Bachmann H., Kok J., unpublished data |
| 14 | MG1614_clu− | Lac +. Transconjugant of MG1614, harbours pLP712 from NCDO712 and show non‐clumping phenotype. | Tarazanova M., Beerthuyzen M., Bachmann H., Kok J., unpublished data |
| 15 | MG1363∆acmA | Derivative of L. lactis MG1363 with deletion of acmA, which leads to chaining phenotype | Steen et al. (2005a) |
| 16 | MG1363∆ahrC | Derivative of L. lactis MG1363 with deletion of ahrC | Larsen et al. (2004) |
| 17 | IL1403∆acmAacmD | Derivative of L. lactis IL1403 with deletion of acmAacmD, which leads to chaining phenotype | Visweswaran et al. (2013) |
| 18 | IL1403(pIL253pil) | EryR; Derivative of L. lactis IL1403 harbouring pilin operon from pSH74 of NCDO712; shows chaining phenotype and high hydrophobicity | Tarazanova et al. (2016) |
| 19 | MG1363∆dltD | Derivative of L. lactis MG1363 with deletion of dltD | Duwat et al. (1997), Steen et al. (2005b) |
| 20 | MG1363(pIL253) | EryR; Derivative of L. lactis MG1363 harbouring plasmid pIL253 | Tarazanova et al. (2016) |
| 21 | IL1403 | Plasmid‐free derivative of L. lactis IL594 | Bolotin et al. (2001) |
| 22 | MG1363(pNZ521;pIL253pil) | CmR and EryR; derivative of L. lactis MG1363 harbouring pNZ521 and pIL253pil (spaCB‐spaA‐srtC1‐srtC2) from the NCDO712 strain | Tarazanova et al. (2016) |
| 23 | MG1363pNZ521 | CmR; derivative of L. lactis MG1363 harbouring proteolytic‐positive genes on pNZ521 | Meijer and Hugenholtz (1997) |
| 24 | MG1363∆galE | Derivative of L. lactis MG1363 with deletion of galE leading to chain formation without galactose in a growth medium | Grossiord et al. (2003) |
| 25 | MG1363pNZ4120 | EryR; derivative of L. lactis MG1363 harbouring eps gene cluster from B40 | Boels et al. (2003) |
| Plasmids | |||
| 1 | pIL253pil | EryR; 13.1 kb; pIL253 harbouring pSH74 pilin operon spaCB‐spaA‐srtC1‐srtC2 with 300 bp upstream region | Tarazanova et al. (2016) |
| 2 | pIL253pilΔ1 | EryR; 11.6 kb; pIL253 harbouring spaCB‐spaA‐srtC1‐srtC2 with 1.5 kb internal deletion in spaCB | Tarazanova et al. (2016) |
| 3 | pNZ521 | CmR; encodes the extracellular serine proteinase (PrtP) from strain SK110 | Meijer and Hugenholtz (1997) |
| 4 | pLP712 | The 55.39 kb plasmid encoding genes for lactose catabolism and a serine proteinase involved in casein degradation | Wegmann et al. (2012) |
| 5 | pIL253 | EryR; 4.9 kb; Low copy‐number derivative of pAMβ1 | Simon and Chopin (1988) |
| 6 | pNZ4120 | EmR; pIL253 derivative containing a 17 kb Ncol fragment carrying the eps gene cluster from NIZO B40 | Boels et al. (2003) |
Table 2.
Phenotypic characteristics of stationary Lactococcus lactis strains measured at pH 6.7. Mean values ± standard deviation of biological triplicates (n = 3) are shown. ZP, zeta potential (mV); CSH, cell surface hydrophobicity (%); NaCN, cell attachment to sodium caseinate (%); NaCN90C, cell attachment to sodium caseinate heated at 90°C for 10 min (%); ParaCN, cell attachment to para‐caseinate (%); E24, emulsion stability after 24 h (%)
| L. lactis | ZP, mV | CSH, % | NaCN, % | NaCN90C, % | ParaCN, % | E24,% |
|---|---|---|---|---|---|---|
| Pili‐overexpressing strains are chaining and clumping | ||||||
| IL1403 | −7.2 ± 0.5 | 0 ± 19.9 | 41.7 ± 13.6 | 67.2 ± 2.2 | 63.7 ± 1.6 | 0 ± 0 |
| IL1403(pIL253pil) | −13.9 ± 1.3* | 93.2 ± 4.5* | 93 ± 2.8* | 94 ± 0.4* | 91.8 ± 1.8* | 88.9 ± 19.3* |
| MG1299 | −30.3 ± 2.1 | 75.5 ± 3.3 | 82.8 ± 2.7 | 82.7 ± 0.3 | 79.2 ± 2.2 | 35.6 ± 24.9 |
| MG1299(pIL253pil) | −18.1 ± 1.3* | 99.1 ± 1.2* | 97.5 ± 1.1* | 99 ± 0.6* | 97 ± 1* | 99.3 ± 0.6 |
| MG1363pIL253 | −26.4 ± 1.8 | 3.8 ± 6.8 | 99.2 ± 0.1 | 99.4 ± 0.3 | 80.6 ± 6.2 | 0 ± 0 |
| MG1363(pIL253pil) | −11.9 ± 0.2* | 94 ± 2.2* | 82.8 ± 7.4 | 81 ± 6.8* | 66 ± 16.4 | 85.2 ± 15* |
| MG1363(pNZ521) | −31.7 ± 0.8 | 18.7 ± 10.1 | 90.3 ± 2.8 | 88.8 ± 2.4 | 79.2 ± 9 | 0 ± 0 |
| MG1363(pNZ521;pIL253pil) | −17 ± 1.9* | 99 ± 1* | 95.4 ± 0.2 | 96.5 ± 2.5 | 89.4 ± 3 | 65.6 ± 29.9* |
| NCDO712 | −20 ± 0.5 | 99.4 ± 0.3 | 96 ± 0.6 | 95.7 ± 2 | 94.7 ± 0.6 | 99.7 ± 0.6 |
| NCDO712(pIL253pil) | −20.5 ± 1.3 | 96.3 ± 0.6 | 85 ± 10.4 | 84.2 ± 9 | 92 ± 3.6 | 100 ± 0 |
| Chaining phenotypeb | ||||||
| IL1403∆acmAacmD | −12 ± 0.4* | 9.5 ± 4.6 | 97.2 ± 0.6* | 96.2 ± 2.1* | 96.4 ± 1.3* | 0 ± 0 |
| MG1363∆acmA | −31.4 ± 1.4 | 15.5 ± 4.5 | 89 ± 0.7* | 87.6 ± 1 | 83.5 ± 10 | 0 ± 0 |
| MG1363∆galE | −28.7 ± 0.8 | 14.9 ± 4.4 | 94.2 ± 2.1* | 95.6 ± 0.5* | 37.5 ± 24.6 | 0 ± 0 |
| MG1363∆dltD | −29.2 ± 0.2 | 15.4 ± 3.5 | 84.9 ± 0.6 | 83.9 ± 3.1 | 82.4 ± 3.2 | 0 ± 0 |
| Non‐chaining, non‐clumping phenotype* | ||||||
| MG1363 | −30.2 ± 0.7 | 5.8 ± 0.2 | 82.3 ± 0.9 | 82.8 ± 1.9 | 79.4 ± 1.7 | 0 ± 0 |
| MG1261 | −30 ± 1.2 | 21.8 ± 0.2* | 93.9 ± 1.4* | 92.6 ± 0.8* | 88.8 ± 6.3 | 0 ± 0 |
| MG1063 | −29 ± 0.9 | 16.7 ± 0* | 89.5 ± 2.6 | 89.6 ± 2.1 | 83 ± 5.1 | 0 ± 0 |
| MG1362 | −31.7 ± 3.3 | 5.6 ± 9.6 | 89.6 ± 3.4 | 84.9 ± 9.7 | 79.6 ± 14.1 | 0 ± 0 |
| MG1365 | −30.3 ± 1.2 | 28.6 ± 6.1* | 92.9 ± 1.8* | 93.1 ± 1.3* | 76.6 ± 26.4 | 0 ± 0 |
| MG1614 | −42 ± 2.4 | 20.4 ± 3.1 | 97.1 ± 0.8 | 94.8 ± 4.1 | 97.7 ± 1.3 | 0 ± 0 |
| MG1614_clu− | −39.4 ± 0.5 | 73.9 ± 4.1* | 96.3 ± 0.9 | 94.6 ± 0.6 | 89.4 ± 4.9 | 0 ± 0 |
| MG1363∆ahrC | −29.5 ± 1.2 | 79.9 ± 10.7* | 84.5 ± 1.1 | 81 ± 2.7 | 80.2 ± 1.7 | 23.8 ± 2.1* |
| MG1363(pIL253pil∆1) | −16.9 ± 0.9* | 79.7 ± 16.9* | 66 ± 11* | 64.5 ± 14.4 | 86.3 ± 5.2 | 97.8 ± 3.9* |
| Clumping phenotypea | ||||||
| MG1614_clu+ | −36 ± 0.4 | 90.2 ± 3.7* | 81.7 ± 15 | 58.1 ± 26.3 | 94.6 ± 0.6 | 31.3 ± 4* |
| EPS producinga | ||||||
| MG1363(pNZ4120) | −18.8 ± 2.2* | 0 ± 23.8 | 96.2 ± 1* | 98.3 ± 0.5* | 97.4 ± 0.2* | 0 ± 0 |
Comparisons were made to L. lactis MG1363 except for MG1614_clu−/clu+ and MG1363(pIL253pil∆1) which were compared with MG1614 and MG1363pIL253 respectively.
Comparisons were made to strains IL1403 and MG1363.
* Significant at P < 0.01 – comparisons were made to non‐surface modified controls.
The 25 strains are variants of the dairy isolate L. lactis ssp. cremoris NCDO712 and its plasmid and phage‐cured derivative MG1363 (Gasson, 1983). Deletion of the genes acmA (Steen et al., 2005a), acmAacmD (Visweswaran et al., 2013), dltD (Steen et al., 2005b) or galE (Grossiord et al., 2003) led to a cell chaining. The introduction of pNZ4120, a plasmid encoding the eps gene cluster from L. lactis NIZO B40 (Boels et al., 2003), in strain MG1363, led to EPS production, which is accompanied by a more positive surface charge (Table 2). We recently isolated strains of L. lactis MG1614 in which the protease/lactose plasmid pLP712 from NCDO712 was introduced through conjugation (Tarazanova M., Beerthuyzen M., Bachmann H., Kok J., unpublished data). As described earlier (Gasson and Davies, 1980), we obtained strains with clumping Clu+ and non‐clumping Clu− phenotypes among the transconjugants carrying pLP712 (Wegmann et al., 2012). Surface characterization of the transconjugants showed that the non‐clumping parent strain MG1614 exhibited a hydrophobicity of ~20 ± 3%, while the hydrophobicity increased to ~74 ± 4% and ~90 ± 4% in the Clu− and Clu+ strains respectively (Table 2).
The strains MG1365, MG1362, MG1063, MG1261 and MG1299 differ from their parent NCDO712 by carrying only one or two plasmids (Gasson, 1983). Although originally described as containing five plasmids, we recently discovered that strain NCDO712 has six plasmids (Tarazanova et al., 2016). One of these plasmids, pSH74, carries a spaCB‐spaA‐srtC1‐srtC2 gene cluster, which encodes proteins that lead to pili formation on the cell surface upon overexpression (Tarazanova et al., 2016). The overexpression of different types of pili in L. lactis is known to cause auto‐aggregation (Oxaran et al., 2012). Our results show that overexpression of spaCB‐spaA‐srtC1‐srtC2 leads to an increase in surface hydrophobicity from 3.8 ± 6.8% to 94 ± 2.2% and a decrease of the cell surface charge from −26.4 ± 1.8 to −11.9 ± 0.2 (Table 2). We also distinguish between chaining and clumping phenotypes. Strains exhibiting a clumping phenotype after mild centrifugation (350 g for 2 min) form strong cell aggregates that remain fast sedimenting clumps after pellet re‐suspension. Such clumping is mainly seen with pili‐overexpressing strains. In contrast, strains with a chaining phenotypes (e.g. strains carrying mutations in acmA, dltD or galE) form long chains of cocci that sediment in overnight cultures. However, after centrifugation the cell pellet can be easily re‐suspended and cells do not clump. Strain MG1363(pIL253pil∆1) contains the spa pilus gene cluster with an internal deletion of 1.5 kb in the spaCB gene encoding the pilin tip protein SpaC and a pilus basal subunit SpaB. This strain forms neither chains nor clumps and retained a high cell surface hydrophobicity. This strain produces pili, but they are not attached to the cell surface (Tarazanova et al., 2016). The used strains differ furthermore in their cell surface charge, emulsification properties and the propensity to bind milk proteins (Table 1). This morphologically diverse collection of strains was used for further analysis.
Acidification rates
As the rate by which L. lactis acidifies the milk during fermentation and the final pH in the dairy product can influence the textural properties of the latter, we followed the development of pH for all strains during 21 h of milk fermentation (Table 3). It is known that very fast acidification of milk can result in excessive syneresis, whereas very slow acidification leads to the formation of a weaker gel (Gastaldi et al., 1997; Lucey, 2004). The maximum acidification rate for most strains was ~0.5 pH/h, but strains showing a chaining phenotype (MG1363∆acmA, MG1363∆galE), the pilin‐decorated strain MG1363(pIL253pil) and its control strain MG1363(pIL253) exhibited slower acidification rates (Table 3). The acidification rates within each comparison group were almost identical, leaving the surface alteration as the main variable for comparisons. The final pH of all milk samples fermented by derivatives of L. lactis MG1363 was 4.25 ± 0.04, with the exception of the proteolytic‐positive pilus‐harbouring strain MG1363(pNZ521;pIL253pil), and strain IL1403 and its derivatives (Table 3), which all exhibited elevated pH values.
Table 3.
Textural properties of milk fermented by surface altered lactococci. Mean values ± standard deviation (n = 3) are shown for independent biological replications for gel hardness (g), viscosity (mPa·s) and pH
| Strain | Gel hardness (g) | pH | Viscosity at shear rate of 10 s−1, mPa·s | Max acidification (n = 1), pH/h |
|---|---|---|---|---|
| Pili‐overexpressing strains and their parental control strains without pili (grouped per comparison) | ||||
| MG1363(pIL253)a | 40.1 ± 2.18 | 4.29 ± 0.05 | 722.7 ± 11.3 | −0.36 |
| MG1363(pIL253pil∆1)a | 45.1 ± 1.13* | 4.23 ± 0 | 1050 ± 19.1* | −0.39 |
| MG1363(pIL253pil)a | 47.8 ± 1.8* | 4.23 ± 0.05 | 1079.3 ± 58.2* | −0.39 |
| MG1363(pNZ521)a | 42.8 ± 0.4 | 4.22 ± 0.05 | 773 ± 64.3 | −0.49 |
| MG1363(pNZ521; pIL253pil)a | 49.1 ± 2.8* | 4.34 ± 0.006 | 1027.2 ± 30* | −0.46 |
| NCDO712a | 43.5 ± 0.6 | 4.24 ± 0.06 | 1498 ± 60.6 | −0.39 |
| NCDO712(pIL253pil)a | 42.4 ± 3.2 | 4.23 ± 0.006 | 1192 ± 146.2* | −0.34 |
| MG1299 | 38.59 ± 0.6 | 4.23 ± 0.05 | 726.1 ± 68.1 | −0.49 |
| MG1299(pIL253pil) | 38.2 ± 0.6 | 4.27 ± 0.001 | 797 ± 54 | −0.54 |
| IL1403 | 37.5 ± 0.64 | 4.45 ± 0.02 | 680.1 ± 34.5 | −0.53 |
| IL1403(pIL253pil) | 37 ± 0.7 | 4.47 ± 0.001 | 745 ± 78 | −0.51 |
| Combinations of EPS and pili/no‐pili forming strains | ||||
| MG1363(pNZ4120)a | 48.9 ± 1.9 | 4.32 ± 0.006 | 4556 ± 37.7* | −0.53 |
| MG1363(pNZ4120) + MG1363(pIL253pil)* | 51.9 ± 1.1 | 4.22 ± 0.006 | 3231 ± 78.9 | NA |
| MG1363(pNZ4120) + MG1363(pIL253pil∆1) | 44.4 ± 3.6 | 4.19 ± 0.006 | 2921 ± 277.7 | NA |
| MG1363(pNZ4120) + MG1363(pIL253)a | 46.9 ± 2.9 | 4.24 ± 0.006 | 3242.7 ± 396.7 | NA |
| MG1363(pNZ4120) + NCDO712 | 45.6 ± 6.8 | 4.18 ± 0.01 | 2847 ± 117.1 | NA |
| MG1363(pNZ4120) + NCDO712(pIL253pil) | 46.97 ± 1.48 | 4.21 ± 0 | 2976 ± 105.5 | NA |
| Chaining strains and their parental controls | ||||
| IL1403 | 37.5 ± 0.64 | 4.45 ± 0.02 | 680.1 ± 34.5 | −0.53 |
| IL1403∆acmAacmD | 35.4 ± 0.14* | 4.39 ± 0.01 | 1008.5 ± 109.3* | −0.51 |
| MG1363 | 43.78 ± 0.89 | 4.24 ± 0.05 | 926.1 ± 50.5 | −0.53 |
| MG1363∆acmA | 41.5 ± 1.08* | 4.24 ± 0.05 | 1034.7 ± 18.9* | −0.42 |
| Transconjugants (clumping/non‐clumping) | ||||
| MG1614 | 40 ± 0.43 | 4.23 ± 0.05 | 832.5 ± 40.9 | −0.54 |
| MG1614_clu− | 39 ± 1.19 | 4.27 ± 0.06 | 739 ± 37* | −0.5 |
| MG1614_clu+ | 41.7 ± 1 | 4.27 ± 0.05 | 887.9 ± 28 | −0.5 |
| NCDO712 derivatives carrying one or two plasmids and MG1363 derivatives with single gene deletions | ||||
| NCDO712 | 42.4 ± 1.3 | 4.24 ± 0.06 | 867.2 ± 106.1 | −0.51 |
| MG1363 | 43.78 ± 0.89 | 4.24 ± 0.05 | 926.1 ± 50.5 | −0.53 |
| MG1363∆ahrC | 43.8 ± 0.43 | 4.28 ± 0.03 | 771 ± 205.5 | −0.56 |
| MG1363∆dltD | 43.9 ± 0.56 | 4.24 ± 0.05 | 891.9 ± 66.9 | −0.52 |
| MG1363∆galE | 44.71 ± 0.58 | 4.21 ± 0.05 | 871.1 ± 181.3 | −0.45 |
| MG1063 | 45.17 ± 0.39 | 4.19 ± 0.05 | 986.97 ± 20.7 | −0.53 |
| MG1261 | 41 ± 0.91 | 4.21 ± 0.05 | 790.97 ± 20.6 | −0.54 |
| MG1362 | 44.1 ± 0.81 | 4.2 ± 0.04 | 945.1 ± 40.9 | −0.52 |
| MG1299 | 38.59 ± 0.6 | 4.23 ± 0.05 | 726.1 ± 68.1 | −0.49 |
NA, not applicable. For cell mixtures, the acidification rate was not determined.
For the indicated strains independent repeats of the experiments on different days confirmed the results with slightly different absolute values.
* Significant at P < 0.05 – comparisons were made to non‐surface modified controls as described in Table 2.
Gel hardness
To determine if cell surface properties influence the rheological parameters of fermented milk, we measured gel hardness and viscosity. The gel hardness of fermented milk prepared with the pili‐overexpressing strain MG1363 (pIL253pil) increased by approximately 46% (P = 0.009) compared with milk fermented with the empty plasmid control strain MG1363(pIL253) (Table 3). Similar to MG1363(pIL253pil), the proteolytic‐positive variant overexpressing pili, strain MG1363(pNZ521; pIL253pil), also increased the fermented milk gel strength, by 15% (P = 0.04). For three other strains, NCDO712(pIL253pil), MG1299(pIL253pil) and IL1403(pIL253pil), gel hardness did not change significantly compared with their control strains. The fact that IL1403(pIL253pil) did not change gel strength can be explained by the very low number of pili on the surface of the cells as compared with MG1363(pIL253pil) (Tarazanova et al., 2016). The data suggest that the overexpression of pili has a profound effect on gel hardness, but the effect seems to be strain‐dependent. When chaining or clumping cells were used gel hardness was significantly decreased. The chain‐forming strains IL1403∆acmAacmD and MG1363∆acmA decreased gel hardness compared with their controls by 5.65% (P = 0.005) and 5.2% (P = 0.049) respectively. Together, these results show that microbial cell surface alterations brought about by either pili overexpression or cell chaining significantly alters the gel hardness of fermented milk.
Viscosity
Viscosity was increased by 19–35% for milk fermented with the strains overexpressing pili, MG1363(pIL253pil) (P = 0.0005) and MG1363(pNZ521; pIL253pil) (P = 0.003), relative to their controls (Table 3). In comparison to milk fermented with the EPS‐producing strain MG1363pNZ4120 (392% increase compared with MG1363, P = 0.0007) the increase in viscosity caused by pili production is considerably lower. We did not observe synergistic effects when milk was fermented with a 1:1 mixture of EPS‐ and pili‐producing strains (Table 3).
The use of chaining phenotypes lead to milk gels with increased viscosity. For example, the chaining strains MG1363∆acmA and IL1403∆acmAacmD formed fermented milk with a viscosity that was 11% (P = 0.025) and ≈ 50% (P = 0.026) higher compared with the control strains MG1363 and IL1403 respectively (Table 3). Overall, the results of milk viscosity measurements show that the engineering of the surface morphology of L. lactis increases product viscosity by up to 50%.
Localization of cells in fermented milk
To investigate if alterations of the bacterial cell surface affect the location of cells in the fermented milk matrix, we performed CLSM imaging on undisturbed fermented milk samples. Due to the sedimentation of cells during fermentation and the limited depth at which CLSM allows imaging (~ 40 μm), the number of cells in the image might be higher than in the average sample. Nevertheless, we consider the observed effects to be indicative for what occurs throughout the product and the CLSM images to provide information on the behaviour of the bacteria in situ. The results (Fig. 1) reveal obvious differences between various strains. For example, loose cocci and clumping cocci locate in the protein matrix close to serum regions (Fig. 1A and C), aggregated cells of MG1363(pIL253pil) fill the serum regions between the aggregated proteins in the fermented milk (Fig. 1B), chaining cells are located in protein and serum regions (Fig. 1D and E), while EPS‐producing cells locate predominantly in serum regions (Fig. 1F). Interestingly, the localization effect observed for MG1363(pIL253pil) was not seen for strain IL1403(pIL253pil). This would be consistent with the fact that pilin overexpression results in much fewer pili on the surface of IL1403 as compared with MG1363 (Tarazanova et al., 2016) and the limited effect on gel hardness presented above. Overall the CLSM results indicate that surface properties of L. lactis can have profound effects on the location of the cells in the fermented milk matrix.
Figure 1.
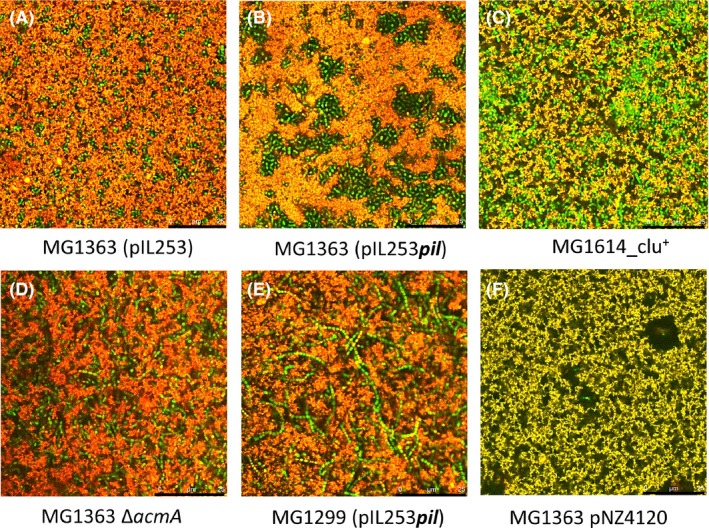
Microstructure of milk fermented by Lactococcus lactis strains with altered surface properties. Bacterial cells are green, while proteins and lipids appear orange/red; the black areas represent the serum fraction. The black bar in the right corner indicates 25 μm.
Discussion
Important physical properties of fermented milk are gel hardness and viscosity. It is known that the hardness of fermented milk increases with a higher dosage of casein (Oliveira et al., 2001), upon heat treatment (Lucey et al., 1997) or homogenization of milk (Serra et al., 2009), decreasing pH (Schkoda et al., 1999), longer acidification time or rate (Kristo et al., 2003) or lower post‐incubation temperature (Lucey and Singh, 1997). The viscosity of fermented milk increases when the gel is allowed to harden before stirring, when stirring intensity is lowered or when the bacterial strains applied produce EPS (Girard and Schaffer‐Lequart, 2007). Here we investigated the potential role of bacterial surface properties and morphology on the textural and structural parameters of fermented milk. Comparisons were made between engineered strains showing alterations in cell chaining, clumping, exopolysaccharide production and pili formation and their isogenic parental strain.
We found that the alterations of cell surface morphology are accompanied by changes in surface charge and hydrophobicity. The bacterial surface properties of these strains also affected the viscosity and gel hardness of milk fermented with them. In addition, the bacterial localization in the fermented milk matrix (Table 4) was dependent on cell surface morphology.
Table 4.
Summary of textural parameters of milk fermented with surface‐altered lactococci
| Phenotype | Straina | Viscosity, % | Gel hardness, % |
|---|---|---|---|
| Pili overexpression | MG1363(pIL253pil) | +49.3** | +19.2** |
| MG1363(pIL253pil∆1) | +45.3* | +12.5* | |
| MG1363(pNZ521; pIL253pil) | +40.1** | +14.7* | |
| MG1299(pIL253pil) | +9.8 | −1 | |
| NCDO712(pIL253pil) | −20.4* | −2.5 | |
| IL1403(pIL253pil) | +9.5 | −1.3 | |
| Chaining | IL1403∆acmAacmD | +48.3* | −5.6** |
| MG1363∆acmA | +11.7* | −5.2* | |
| Clumping | MG1614_clu+ | +6.7 | +4.3 |
All comparisons are made with the isogenic parent strain.
* Significance (P < 0.05).
** Significance (P < 0.01).
The bacterial cell surface contains charged hydrophilic as well as hydrophobic areas, which are the result of the complex molecular composition of the cell wall. For example, overexpression of pili in MG1363(pIL253pil) led, in neutral pH, to a decrease in charge (from −26 ± 2 mV to −12 ± 0 mV). The decrease in net‐negative charge can possibly be explained by the slightly net‐positive charge of the overexpressed pilin proteins. The charged residues of pilin proteins are likely to be on the outside of the pilus (Tsumoto et al., 2004). Analysis of the spa pilin protein sequences (http://www.expasy.org/) showed 30–49% hydrophobic residues. The increase in hydrophobicity from 5% to 96% of MG1363(pIL253pil) relative to its parent might be explained by hydrophobic patches on the overexpressed pili. Taken together bacterial amphiphilic surface properties are governed by the molecular composition of its cell wall, which is strongly strain‐dependent (Tarazanova et al., 2017) and can be engineered.
The increased viscosity and gel hardness of fermented milk using pili overexpression strains was seen independently for three out of the six strains tested, which indicates that this effect is strain‐dependent. The three strains where this effect was not observed are NCDO712, MG1299 and IL1403. Strain NCDO712 carries the spa pilin genes on the low copy‐number plasmid pSH74, which are not expressed at high levels (Tarazanova et al., 2016). Similarly, electron microscopy revealed that IL1403(pIL253pil) carried low numbers of pili on the surface (Tarazanova et al., 2016). Strain MG1299(pIL253pil) forming long chains shows a trend towards higher viscosity but it has no effect on gel hardness. This phenomenon is in agreement with our observations for solely chaining strains described below. The 20–35% increase in viscosity obtained with the other strains is a substantial alteration that is likely to be perceived sensorically.
The effect of the microbial surface on textural properties of fermented milk leads us to hypothesize that bacteria can be seen and used as structural or ingredient elements of a food matrix. As such they engage in physico‐chemical interactions with milk proteins and/or with other cells via molecules on their polymeric cell surface layers (Ubbink and Schär‐Zammaretti, 2005; Jacobs et al., 2007; Burgain et al., 2013). The occurrence of interactions is a balance between attractive and repulsive forces, which strongly depend on the interacting particles. For instance, it was shown that for Lactobacillus bulgaricus GG the force between micellar casein (or denatured whey protein) and the bacterial cell is about 0.4 nN, but this was significantly different for other strains (Burgain et al., 2014b). Together with our results, these observations suggest that the type and force of microbe–matrix interactions can influence the structure, stability and textural properties of the fermented milk matrix.
Strains of L. lactis show a high degree of biodiversity in surface properties among which their capacity to bind to milk proteins. We have previously reported that dairy isolates display a higher milk protein binding affinity than plant isolates (Tarazanova et al., 2017). The increased viscosity and gel hardness of fermented milk might, thus, also be caused by bacterial interactions with milk proteins (Tarazanova et al., 2017) as well as by cell to cell connections via pili between the starter culture cells when they are in a close proximity (Tarazanova et al., 2016). The results of the current study show that despite the surface alterations inflicted, the lactococcal cells retained a high capacity to bind to milk proteins (Table 2). These results indicate that factors other than pili or morphology changes are responsible for milk protein binding.
Interactions of L. lactis with milk proteins might result in preferential localization of the bacterial cells in the protein matrix or in serum regions, in the form of aggregates and/or chains. This is corroborated by microscopic observations of pili‐overexpressing cells in the milk matrix. These cells seem to be localized in the serum regions (Fig. 1). Here the size and shape of particles is of importance. Cells do not only form very strong cell–cell connections, leading to cell aggregates located in milk gel cavities, but also cell–protein contacts between pili of outer cells of aggregates and proteins of milk gel, which appeared to contribute to the increased milk gel hardness.
Similar to pili‐expressing cells, which show a chaining and, after centrifugation, a clumping phenotype, bacteria that only form chains (acmA/acmD deletions) increased the viscosity up to 12–48%, and in contrast to pili‐expressing cells this lead to a decrease in gel hardness by 6–14% (Tables 2 and 3). It was detected that there are more cavities in milk gel fermented with chaining strains. The decreased gel hardness can be explained by cavities of serum in the milk matrix: they do not provide any additional bonds to strengthen the gel. We speculate that that the cells do not interact with the aggregated casein micelles in this case and are, thus, not included in the matrix but act like structure breakers or inert fillers. This would be similar to what is seen for fat droplets with natural milk fat globular membranes in milk gels made with unhomogenized milk (Lucey and Singh, 1997): in this case, the matrix has to form around those droplets. Also longer cell chains can be considered as “viscosifying” molecules and not as enhancing properties for the gel hardness. Chain length might be at the basis of the increased viscosity of a matrix obtained by fermenting milk with chaining cells. In general, the longer the molecule (especially when it is stretched) the more viscosifying properties it has due to possible hydrogen bonding with water molecules as well as molecule–molecule interactions or entanglements.
The findings presented here show that engineering surface properties of dairy starter strains allows altering product properties such as gel hardness and milk viscosity. It will be highly interesting to see whether the quality changes can be perceived in sensory analyses. This opens possibilities to develop new concepts in improving fermented products with altered textural properties.
Experimental procedures
Bacterial strains and growth conditions
Lactococcus lactis strains (Table 1) were grown at 30°C in M17 broth (Oxoid Ltd, Basingstoke, UK) containing 1% glucose (GM17) or 1% lactose (LM17). When required, erythromycin (Ery; 10 μg ml−1), chloramphenicol (Cm; 5 μg ml−1), rifampicin (Rif; 50 μg ml−1) or streptomycin (Str; 100 μg ml−1) were added to the indicated final concentrations.
Cell surface properties
Bacterial cell surface properties, i.e. surface charge (or zeta potential, ZP, mV), cell surface hydrophobicity (CSH, %), emulsion stability after 24 h (E24, %) and attachment to milk proteins were measured as described earlier (Tarazanova et al., 2017).
Preparation of fermented milk
Full‐fat pasteurized and homogenized milk (3.6% fat, 3.5% protein, 4.7% lactose) was purchased from a local supplier and sterilized at 115°C for 15 min to denature the whey proteins (Sodini et al., 2004). Sterilized milk was supplemented with sterile 50% glucose solution (4% final concentration) and sterile 20% Bacto™ Casitone (Pancreatic digest of casein, BD, Sparks, MD, USA) solution to 0.2% final concentration to ensure growth of lactose‐negative and protease‐negative strains respectively. Instead, for lactose‐positive and protease‐positive strains, a same volume of water was added to the milk to ensure that protein and fat content of the milk were the same for all strains. For gel strength and viscosity measurements on the fermented milk samples, the bacterial strains were pre‐cultured overnight in milk supplemented with the appropriate antibiotic, but the antibiotic was not added during the actual milk fermentation.
Warm (30°C) milk was inoculated with 2% v/v of an overnight culture. Acidification rates and final pH were measured with a Cinac 14 ph+2T (Alliance instruments, Freppilon, France). pH electrodes were inserted into the inoculated milk samples in 10 ml tubes and measurement was done every 6 min for 21 h at 30°C.
Gel strength of fermented milk
Aliquots (300 ml) of milk without antibiotic were inoculated with 2% of the strains to be tested. The prepared milk was distributed to three 100 ml sterile glass cups (70 mm diameter), which were incubated statically for 21 h at 30°C. Gel strength was measured with a Texture Analyzer (TA.XTplus, Stable Micro Systems Ltd., Sprundel, NL) equipped with a 5 kg load cell according to the manufacture application guidelines. The 100 ml fermented milk samples were compressed uniaxially to a depth of 20 mm with a constant speed of 1 mm s−1 by a probe with a grid‐like geometry having 10 mm side squared openings. The peak force applied on the sample corresponds to the hardness of the milk gel.
Viscosity of fermented milk
After the texture analysis, the viscosity of the fermented milk was measured with a rotational viscometer (Haake Searle RV20 Rotovisco and RC 20 Rheocontroller, ThermoScientific, Hofheim, Germany) with MV2P (medium viscous profile) rotor according to the manufacture guidelines. Fermented milk (60 g) was transferred to the MVP cup and allowed to rest for 15 min. Subsequently, the sample was measured as the shear rate gradually increased from 0 to 400 s−1 for 5 min and then decreased again from 400 to 0 s−1 for 5 min at 21°C. Viscosity values were determined at a shear rate of 10 s−1 of the first curve, i.e. when shear rate was increasing.
Confocal laser scanning microscopy (CLSM)
To 1 ml of homogenized sterilized milk, with sugar and antibiotics as required, 15 μl of a solution containing 0.5% (final concentration) Acridine Orange (Sigma‐Aldrich Chemie B.V., Zwijndrecht, The Netherlands) and 0.025% (final concentration) Rhodamine B (Sigma‐Aldrich Chemie B.V., Zwijndrecht, The Netherlands) in water was added to stain proteins and fat droplets respectively. This milk was inoculated with 1% of an overnight culture, after which 1 μl Syto 9 (5 mM in DMSO, ThermoFisher; 1 μl/OD600 of 1/ml) was immediately added to the sample to stain the bacterial cells. The milk sample was subsequently transferred to a CLSM slide with a cylindrical plastic cup attached to it which was then covered with a lid (Hassan et al., 1995) to prevent evaporation. The sample was incubated for 21 h at 30°C.
CLSM images were taken using a Leica TCS SP 5 confocal laser scanning microscope (Leica Microsystems CMS GmbH, Mannheim, Germany) with Leica application Suite Advanced Fluorescence v. 2.7.3. build. 9723. The Argon laser was used to visualize the Syto9‐stained bacteria, while the DPSS 561 laser was used to visualize the milk protein and fat droplet in the matrix that were stained by the Acridine Orange and Rhodamine B respectively. The objective lens used was an HCX PL APO 63 × /1.2/water CORR CS.
Conflict of interests
The project was funded by TiFN, a public–private partnership on precompetitive research in food and nutrition. The public partners are responsible for the study design, data collection and analysis, decision to publish and preparation of the manuscript. The private partners have contributed to the project through regular discussion. A patent application pertaining to the presented findings was filed.
Acknowledgements
We thank Venera Proneva, Jan Klok and Saskia de Jong for technical assistance. Furthermore, we thank Fred van de Velde for valuable discussions and suggestions.
Microbial Biotechnology (2018) 11(4), 770–780
Funding information
The project was funded by TiFN, a public–private partnership on precompetitive research in food and nutrition.
References
- Amatayakul, T. , Halmos, A.L. , Sherkat, F. , and Shah, N.P. (2006) Physical characteristics of yoghurts made using exopolysaccharide‐producing starter cultures and varying casein to whey protein ratios. Int Dairy J 16: 40–51. [Google Scholar]
- Boels, I.C. , Van Kranenburg, R. , Kanning, M.W. , Chong, B.F. , De Vos, W.M. , and Kleerebezem, M. (2003) Increased exopolysaccharide production in Lactococcus lactis due to increased levels of expression of the NIZO B40 eps gene cluster. Appl Environ Microbiol 69: 5029–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin, A. , Wincker, P. , Mauger, S. , Jaillon, O. , Malarme, K. , Weissenbach, J. , et al (2001) The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11: 731–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosshard, H.R. , Marti, D.N. , and Jelesarov, I. (2004) Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J Mol Recognit 17: 1–16. [DOI] [PubMed] [Google Scholar]
- Broadbent, J.R. , and Kondo, J.K. (1991) Genetic construction of nisin‐producing Lactococcus lactis subsp. cremoris and analysis of a rapid method for conjugation. Appl Environ Microbiol 57: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist, G. , Karsens, H. , Nauta, A. , van Sinderen, D. , Venema, G. , and Kok, J. (1997) Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl Environ Microbiol 63: 2722–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgain, J. , Gaiani, C. , Francius, G. , Revol‐Junelles, A.M. , Cailliez‐Grimal, C. , Lebeer, S. , et al (2013) In vitro interactions between probiotic bacteria and milk proteins probed by atomic force microscopy. Colloids Surf B Biointerfaces 104: 153–162. [DOI] [PubMed] [Google Scholar]
- Burgain, J. , Scher, J. , Francius, G. , Borges, F. , Corgneau, M. , Revol‐Junelles, A.M. , et al (2014a) Lactic acid bacteria in dairy food: surface characterization and interactions with food matrix components. Adv Colloid Interface Sci 213: 21–35. [DOI] [PubMed] [Google Scholar]
- Burgain, J. , Scher, J. , Lebeer, S. , Vanderleyden, J. , Cailliez‐Grimal, C. , Corgneau, M. , et al (2014b) Significance of bacterial surface molecules interactions with milk proteins to enhance microencapsulation of Lactobacillus rhamnosus GG. Food Hydrocoll 41: 60–70. [Google Scholar]
- Burgain, J. , Scher, J. , Lebeer, S. , Vanderleyden, J. , Corgneau, M. , Guerin, J. , et al (2015) Impacts of pH‐mediated EPS structure on probiotic bacterial pili–whey proteins interactions. Colloids Surf B Biointerfaces 134: 332–338. [DOI] [PubMed] [Google Scholar]
- Busscher, H. , and Weerkamp, A. (1987) Specific and non‐specific interactions in bacterial adhesion to solid substrata. FEMS Microbiol Lett 46: 165–173. [Google Scholar]
- Cheng, J. , Ma, Y. , Li, X. , Yan, T. , and Cui, J. (2015) Effects of milk protein‐polysaccharide interactions on the stability of ice cream mix model systems. Food Hydrocoll 45: 327–336. [Google Scholar]
- Delcour, J. , Ferain, T. , Deghorain, M. , Palumbo, E. , and Hols, P. (1999) The biosynthesis and functionality of the cell‐wall of lactic acid bacteria. Antonie Van Leeuwenhoek 76: 159–184. [PubMed] [Google Scholar]
- Dickinson, E. (2015) Microgels – an alternative colloidal ingredient for stabilization of food emulsions. Trends Food Sci Technol 43: 178–188. [Google Scholar]
- Duwat, P. , Cochu, A. , Ehrlich, S.D. , and Gruss, A. (1997) Characterization of Lactococcus lactis UV‐sensitive mutants obtained by ISS1 transposition. J Bacteriol 179: 4473–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, M. , Ratcliffe, I. , and Williams, P.A. (2013) Emulsion stabilisation using polysaccharide–protein complexes. Curr Opin Colloid Interface Sci 18: 272–282. [Google Scholar]
- Gasson, M.J. (1983) Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast‐induced curing. J Bacteriol 154: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson, M.J. , and Davies, F.L. (1980) High‐frequency conjugation associated with Streptococcus lactis donor cell aggregation. J Bacteriol 143: 1260–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson, M.J. , Godon, J.J. , Pillidge, C.J. , Eaton, T.J. , Jury, K. , and Shearman, C.A. (1995) Characterization and exploitation of conjugation in Lactococcus lactis . Int Dairy J 5: 757–762. [Google Scholar]
- Gastaldi, E. , Lagaude, A. , Marchesseau, S. , and Tarodo de la Fuente, B. (1997) Acid milk gel formation as affected by total solids content. J Food Sci 62: 671–687. [Google Scholar]
- Giaouris, E. , Chapot‐Chartier, M.‐P. , and Briandet, R. (2009) Surface physicochemical analysis of natural Lactococcus lactis strains reveals the existence of hydrophobic and low charged strains with altered adhesive properties. Int J Food Microbiol 131: 2–9. [DOI] [PubMed] [Google Scholar]
- Girard, M. , and Schaffer‐Lequart, C. (2007) Gelation and resistance to shearing of fermented milk: role of exopolysaccharides. Int Dairy J 17: 666–673. [Google Scholar]
- Grossiord, B.P. , Luesink, E.J. , Vaughan, E.E. , Arnaud, A. , and de Vos, W.M. (2003) Characterization, expression, and mutation of the Lactococcus lactis galPMKTE genes, involved in galactose utilization via the Leloir pathway. J Bacteriol 185: 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habimana, O. , Le Goff, C. , Juillard, V. , Bellon‐Fontaine, M.‐N. , Buist, G. , Kulakauskas, S. , and Briandet, R. (2007) Positive role of cell wall anchored proteinase PrtP in adhesion of lactococci. BMC Microbiol 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadde, E.K. , Nicholson, T.M. , Cichero, J.A.Y. , and Deblauwe, C. (2015) Rheological characterisation of thickened milk components (protein, lactose and minerals). J Food Eng 166: 263–267. [Google Scholar]
- Hassan, A.N. , Frank, J.F. , Farmer, M.A. , Schmidt, K.A. , and Shalabi, S.I. (1995) Formation of yogurt microstructure and three‐dimensional visualization as determined by confocal scanning laser microscopy. J Dairy Sci 78: 2629–2636. [DOI] [PubMed] [Google Scholar]
- Hassan, A.N. , Ipsen, R. , Janzen, T. , and Qvist, K.B. (2003) Microstructure and rheology of yogurt made with cultures differing only in their ability to produce exopolysaccharides. J Dairy Sci 86: 1632–1638. [DOI] [PubMed] [Google Scholar]
- Jacobs, A. , Lafolie, F. , Herry, J.M. , and Debroux, M. (2007) Kinetic adhesion of bacterial cells to sand: cell surface properties and adhesion rate. Colloids Surf B Biointerfaces 59: 35–45. [DOI] [PubMed] [Google Scholar]
- Jarunglumlert, T. , Nakagawa, K. , and Adachi, S. (2015) Influence of aggregate structure of casein on the encapsulation efficiency of β‐carotene entrapped via hydrophobic interaction. Food Struct 5: 42–50. [Google Scholar]
- Jeanson, S. , Floury, J. , Gagnaire, V. , Lortal, S. , and Thierry, A. (2015) Bacterial colonies in solid media and foods: a review on their growth and interactions with the micro‐environment. Front Microbiol 6: 1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelen, P. (2005) Advanced dairy chemistry. Int Dairy J 15: 189. [Google Scholar]
- Joyner (Melito), H.S. , and Damiano, H. (2015) Influence of various hydrocolloids on cottage cheese cream dressing stability. Int Dairy J 51: 24–33. [Google Scholar]
- Kristo, E. , Biliaderis, C.G. , and Tzanetakis, N. (2003) Modelling of rheological, microbiological and acidification properties of a fermented milk product containing a probiotic strain of Lactobacillus paracasei . Int Dairy J 13: 517–528. [Google Scholar]
- Larsen, R. , Buist, G. , Kuipers, O.P. , and Kok, J. (2004) ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis . J Bacteriol 186: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Boucher, C. , Gagnaire, V. , Briard‐Bion, V. , Jardin, J. , Maillard, M.‐B. , Dervilly‐Pinel, G. , et al (2016) Spatial distribution of Lactococcus lactis colonies modulates the production of major metabolites during the ripening of a model cheese. Appl Environ Microbiol 82: 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy, F. , and De Vuyst, L. (2004) Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol 15: 67–78. [Google Scholar]
- Li, J.‐M. , and Nie, S.‐P. (2015) The functional and nutritional aspects of hydrocolloids in foods. Food Hydrocoll 53: 46–61. [Google Scholar]
- Lucey, J.A. (2004) Cultured dairy products: an overview of their gelation and texture properties. Int J Dairy Technol 57: 77–84. [Google Scholar]
- Lucey, J.A. , and Singh, H. (1997) Formation and physical properties of acid milk gels: a review. Food Res Int 30: 529–542. [Google Scholar]
- Lucey, J.A. , Teo, C.T. , Munro, P.A. , and Singh, H. (1997) Rheological properties at small (dynamic) and large (yield) deformations of acid gels made from heated milk. J Dairy Res 64: 591–600. [Google Scholar]
- Lucey, J.A. , Otter, D. , and Horne, D.S. (2017) A 100‐year review: progress on the chemistry of milk and its components. J Dairy Sci 100: 9916–9932. [DOI] [PubMed] [Google Scholar]
- Ly, M. , Covarrubias‐Cervantes, M. , Dury‐Brun, C. , Bordet, S. , Voilley, A. , Le, T. , et al (2008a) Retention of aroma compounds by lactic acid bacteria in model food media. Food Hydrocoll 22: 211–217. [Google Scholar]
- Ly, M.H. , Aguedo, M. , Goudot, S. , Le, M.L. , Cayot, P. , Teixeira, J.A. , et al (2008b) Interactions between bacterial surfaces and milk proteins, impact on food emulsions stability. Food Hydrocoll 22: 742–751. [Google Scholar]
- Ly‐Chatain, M.H. , Linh, M. , Le, M.L. , Belin, J. , Waché, Y. , Thanh, M.Le. , et al (2010) Cell surface properties affect colonisation of raw milk by lactic acid bacteria at the microstructure level. Food Res Int 43: 1594–1602. [Google Scholar]
- Mao, Y. , and McClements, D.J. (2013) Modulation of food texture using controlled heteroaggregation of lipid droplets: principles and applications. J Appl Polym Sci 130: 3833–3841. [Google Scholar]
- Meijer, W.C. , and Hugenholtz, J. (1997) Proteolytic enzyme activity in lactococci grown in different pretreated milk media. J Appl Microbiol 83: 139–146. [DOI] [PubMed] [Google Scholar]
- Meyrand, M. , Guillot, A. , Goin, M. , Furlan, S. , Armalyte, J. , Kulakauskas, S. , et al (2013) Surface proteome analysis of a natural isolate of Lactococcus lactis reveals the presence of pili able to bind human intestinal epithelial cells. Mol Cell Proteomics 12: 3935–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell, P. , Fiszman, S.M. , Varela, P. , and Hernando, I. (2014) Hydrocolloids for enhancing satiety: relating oral digestion to rheology, structure and sensory perception. Food Hydrocoll 41: 343–353. [Google Scholar]
- Neve, H. , Geis, A. , and Teuber, M. (1987) Conjugation, a common plasmid transfer mechanism in lactic acid streptococci of dairy starter cultures. Syst Appl Microbiol 9: 151–157. [Google Scholar]
- Oliveira, M.N. , Sodini, I. , Remeuf, F. , and Corrieu, G. (2001) Effect of milk supplementation and culture composition on acidification, textural properties and microbiological stability of fermented milks containing probiotic bacteria. Int Dairy J 11: 935–942. [Google Scholar]
- Oxaran, V. , Ledue‐Clier, F. , Dieye, Y. , Herry, J.‐M. , Péchoux, C. , Meylheuc, T. , et al (2012) Pilus biogenesis in Lactococcus lactis: molecular characterization and role in aggregation and biofilm formation. PLoS ONE 7: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploux, L. , Ponche, A. , and Anselme, K. (2010) Bacteria/material interfaces: role of the material and cell wall properties. J Adhes Sci Technol 24: 2165–2201. [Google Scholar]
- Prasanna, P.H.P. , Grandison, A.S. , and Charalampopoulos, D. (2013) Microbiological, chemical and rheological properties of low fat set yoghurt produced with exopolysaccharide (EPS) producing Bifidobacterium strains. Food Res Int 51: 15–22. [Google Scholar]
- Schkoda, P. , Hechler, A. , and Kessler, H.G. (1999) Effect of minerals and pH on rheological properties and syneresis of milk‐based acid gels. Int Dairy J 9: 269–273. [Google Scholar]
- Serra, M. , Trujillo, A.J. , Guamis, B. , and Ferragut, V. (2009) Evaluation of physical properties during storage of set and stirred yogurts made from ultra‐high pressure homogenization‐treated milk. Food Hydrocoll 23: 82–91. [Google Scholar]
- Simon, D. , and Chopin, A. (1988) Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis . Biochimie 70: 559–566. [DOI] [PubMed] [Google Scholar]
- Smid, E.J. , and Kleerebezem, M. (2014) Production of aroma compounds in lactic fermentations. Annu Rev Food Sci Technol 5: 313–326. [DOI] [PubMed] [Google Scholar]
- Smit, B.A. , Engels, W.J.M. , Alewijn, M. , Lommerse, G.T.C.A. , Kippersluijs, E.A.H. , Wouters, J.T.M. , and Smit, G. (2004) Chemical conversion of alpha‐keto acids in relation to flavor formation in fermented foods. J Agric Food Chem 52: 1263–1268. [DOI] [PubMed] [Google Scholar]
- Smit, B.A. , van Hylckama Vlieg, J.E.T. , Engels, W.J.M. , Meijer, L. , Wouters, J.T.M. , and Smit, G. (2005a) Identification, cloning, and characterization of a Lactococcus lactis branched‐chain alpha‐keto acid decarboxylase involved in flavor formation. Appl Environ Microbiol 71: 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, G. , Smit, B. , and Engels, W. (2005b) Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev 29: 591–610. [DOI] [PubMed] [Google Scholar]
- Sodini, I. , Remeuf, F. , Haddad, S. , and Corrieu, G. (2004) The relative effect of milk base, starter, and process on yogurt texture: a review. Crit Rev Food Sci Nutr 44: 113–137. [DOI] [PubMed] [Google Scholar]
- Steen, A. , Buist, G. , Horsburgh, G.J. , Venema, G. , Kuipers, O.P. , Foster, S.J. , and Kok, J. (2005a) AcmA of Lactococcus lactis is an N‐acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J 272: 2854–2868. [DOI] [PubMed] [Google Scholar]
- Steen, A. , Palumbo, E. , Deghorain, M. , Cocconcelli, P.S. , Delcour, J. , Kuipers, O.P. , et al (2005b) Autolysis of Lactococcus lactis is increased upon D‐alanine depletion of peptidoglycan and lipoteichoic acids. J Bacteriol 187: 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles, M.E. , and Holzapfel, W.H. (1997) Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol 36: 1–29. [DOI] [PubMed] [Google Scholar]
- Tarazanova, M. , Beerthuyzen, M. , Siezen, R. , Fernandez‐Gutierrez, M.M. , de Jong, A. , van der Meulen, S. , et al (2016) Plasmid complement of Lactococcus lactis NCDO712 reveals a novel pilus gene cluster. PLoS ONE 11: e0167970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazanova, M. , Huppertz, T. , Beerthuyzen, M. , van Schalkwijk, S. , Janssen, P. , Wels, M. , et al (2017) Cell surface properties of Lactococcus lactis reveal milk protein binding specifically evolved in dairy isolates. Front Microbiol 8: 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumoto, K. , Umetsu, M. , Kumagai, I. , Ejima, D. , Philo, J.S. , and Arakawa, T. (2004) Role of arginine in protein refolding, solubilization, and purification. Biotechnol Prog 20: 1301–1308. [DOI] [PubMed] [Google Scholar]
- Tuinier, R. , van Casteren, W.H.M. , Looijesteijn, P.J. , Schols, H.A. , Voragen, A.G.J. , and Zoon, P. (2001) Effects of structural modifications on some physical characteristics of exopolysaccharides from Lactococcus lactis . Biopolymers 59: 160–166. [DOI] [PubMed] [Google Scholar]
- Tuinier, R. , Fan, T.‐H. , and Taniguchi, T. (2015) Depletion and the dynamics in colloid–polymer mixtures. Curr Opin Colloid Interface Sci 20: 66–70. [Google Scholar]
- Ubbink, J. , and Schär‐Zammaretti, P. (2005) Probing bacterial interactions: integrated approaches combining atomic force microscopy, electron microscopy and biophysical techniques. Micron 36: 293–320. [DOI] [PubMed] [Google Scholar]
- Visweswaran, G.R.R. , Steen, A. , Leenhouts, K. , Szeliga, M. , Ruban, B. , Hesseling‐Meinders, A. , et al (2013) AcmD, a homolog of the major autolysin AcmA of Lactococcus lactis, binds to the cell wall and contributes to cell separation and autolysis. PLoS ONE 8: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Cao, Y. , Zhang, K. , Fang, Y. , Nishinari, K. , and Phillips, G.O. (2015) Hydrogen bonding enhances the electrostatic complex coacervation between κ‐carrageenan and gelatin. Colloids Surf A Physicochem Eng Aspects 482: 604–610. [Google Scholar]
- Wegmann, U. , Overweg, K. , Jeanson, S. , Gasson, M. , and Shearman, C. (2012) Molecular characterization and structural instability of the industrially important composite metabolic plasmid pLP712. Microbiology 158: 2936–2945. [DOI] [PubMed] [Google Scholar]


