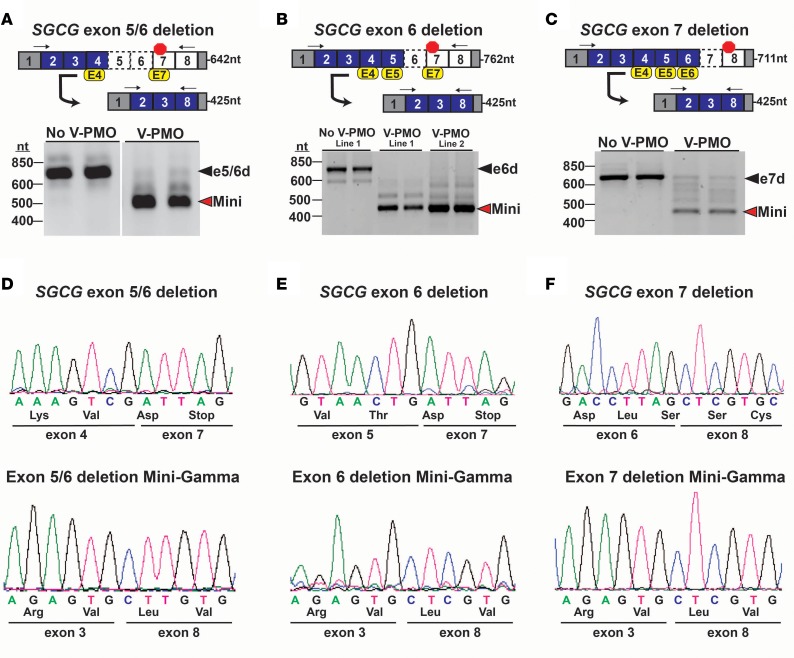
Figure 5. Reading frame correction of multiple SGCG frameshift mutations.
(A–C) Top: Mutation-specific multiexon-skipping strategies were designed to correct the SGCG reading frame of multiple unrelated LGMD 2C patients whose primary SGCG mutations include (A) the deletion of exons 5 and 6; (B) the deletion of exon 6; and (C) the deletion of exon 7. Myogenic cells were treated with vivo-PMO exon skipping cocktails. Arrows denote the location of the RT-PCR primers, and the expected amplicon lengths are indicated. (A–C) Bottom: RT-PCR analysis confirmed the mutant SGCG transcripts were expressed in the reprogrammed cells at the expected size, consistent with their exon deletion status (black arrowheads). Cells treated with the mutation-specific vivo-PMOs cocktails demonstrated predominate expression of the reading frame–corrected Mini-Gamma transcript (red arrowheads). (D–F) Sequence analysis of RT-PCR products: (D) a deletion of exons 5 and 6; (E) a deletion of exon 6; and (F) a deletion of exon 7. Sequence analysis also confirmed the generation of the Mini-Gamma transcript in each of the mutant cell lines, with the splicing of SGCG exons 3 and 8 demonstrating the removal of (D) exons 4 and 7; (E) exon 4, 5, and 7; and (F) exons 4, 5 and 6 from the mature transcripts.