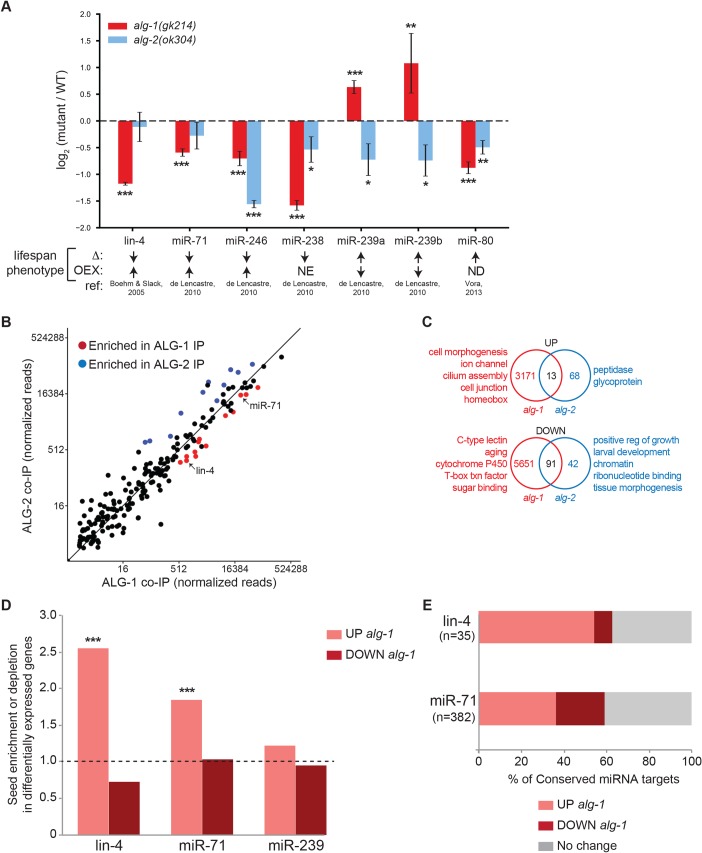
Fig 4. Altered miRNA and mRNA expression in alg-1 and alg-2 mutants.
(A) TaqMan analyses of the indicated miRNAs in alg-1(gk214) (red) and alg-2(ok304) (blue) compared to WT animals at day 5 of adulthood, averaged from five independent experiments. The error bars represent SEMs. *P<0.05, **P<0.01, ***P<0.001 (t-test). The published lifespan phenotypes observed with reduced expression (Δ), overexpression (OEX), and corresponding references (ref) are indicated. ↑ (increased lifespan), ↓ (decreased lifespan), NE (no effect), ND (not determined). (B) Enrichment of specific miRNAs with ALG-1 (red) or ALG-2 (blue) detected by sequencing of small RNAs that co-immunoprecipitated (co-IP) with each AGO protein at day 5 of adulthood averaged from 2 independent experiments. See S2 Table for a complete list of miRNAs reproducibly enriched for association with ALG-1 or ALG-2. (C) Overlap of all genes significantly (P<0.05) up- or down-regulated at day 5 of adulthood in alg-1(gk214) (red) and alg-2(ok304) (blue) compared to WT animals. Overlap of down-regulated genes in alg-1(gk214) and alg-2(ok304) is more than expected by chance (P<0.0001, hypergeometric test). The top ranked gene ontology terms identified by DAVID analysis are listed. See S3 Table and S4 Table for complete RNA-seq and DAVID results. (D) Enrichment and depletion of seed pairing (nucleotides 2–7) for the indicated miRNAs with genes differentially expressed in alg-1(gk214) versus WT animals. The fold difference shown is in comparison to the fraction of seed-pairing sites detected in genes with unchanged expression patterns in the alg-1 mutants. ***P<0.0001 (Chi Squared with Yates Correction). (E) Percent of conserved lin-4 and miR-71 targets predicted by TargetScan [36, 37] that are differentially regulated in alg-1(gk214).