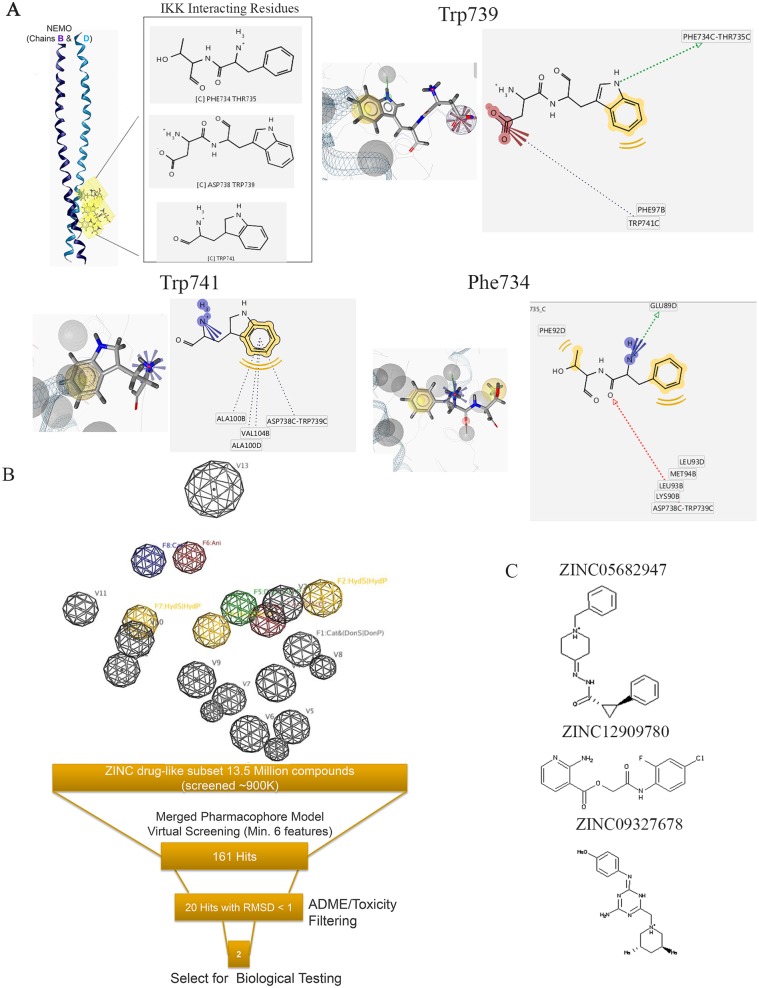
Fig 1. The development of a structure-based pharmacophore model for in silico screen of NBD mimetics.
(A) Interacting residues extracted from the X-ray structure of the NEMO/IKKβ complex (PDB ID: 3BRV) used in the generation of the structure-based pharmacophore model [21]. (B) A 3D representation of the structure-based pharmacophore model. Three hydrophobic groups (F2, F4, and F7: yellow spheres), 1 hydrogen-bond acceptor features (F3: red sphere), 1 hydrogen-bond donor (F5: green sphere), 1 positive ionizable area (F8: blue sphere), 1 negative ionizable (F6: red sphere), and 13 excluded volumes (gray spheres) are shown. (C) Chemical structures of 3 compounds that entered biological testing [24]. 3D, three-dimensional; IKK, IκB kinase; NBD, NEMO-binding domain; NEMO, NF-κB essential modulator; PDB, Protein Data Bank.