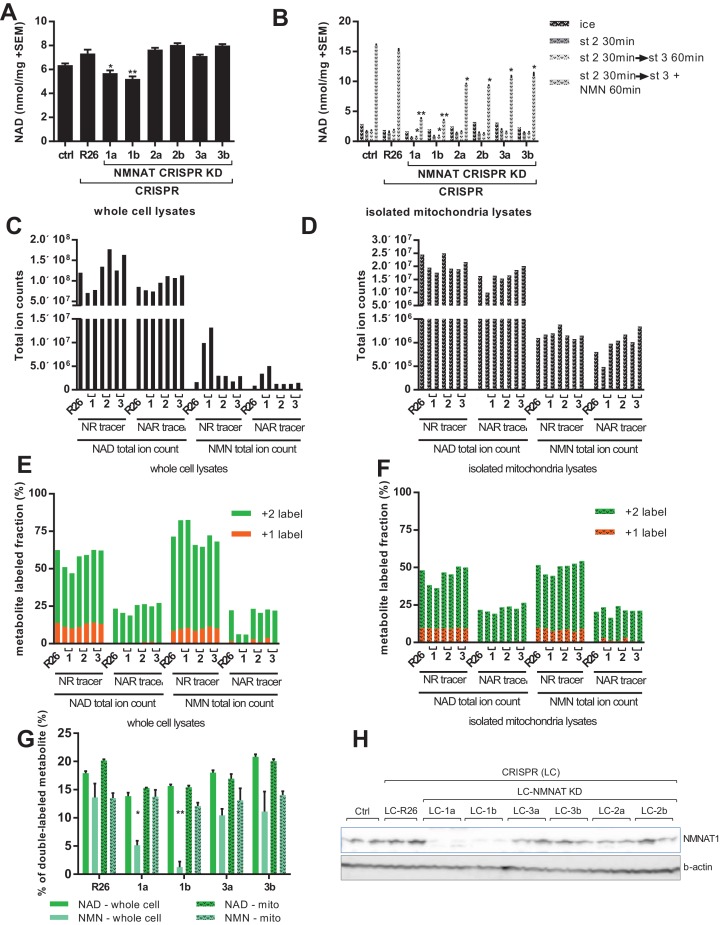
Figure 6. Labeling of mitochondrial NAD tracks that of total NAD, but not of total NMN.
For all panels, data representing whole cells are depicted as solid bars, whereas data from isolated mitochondria are shown with a stippled pattern. (A) Differentiated C2C12 parental and LentiCRISPR transgenic myotubes were analyzed for NAD content. The cells are as follows: ctrl- parental line with no vector; R26- vector control; 1a and 1b- two separate guide RNAs targeting NMNAT1; 2a and 2b- two separate guide RNAs targeting NMNAT2; 3a and 3b- two separate guide RNAs targeting NMNAT3. (N = 3). (B) Mitochondria isolated from differentiated C2C12 cells were held in state 2 (MirO5 respiration buffer containing 10 mM Pyruvate, 5 mM Malate) at 37°C with shaking for 30 min. They were then collected and lysed in perchloric acid immediately, or transitioned into state three by adding ADP (12.5 mM, final concentration) with or without supplementation with NMN (0.5 mM, final concentration) and maintained for 60 min at 37°C with shaking before collection. (N = 2–4). (C–D) Total ion counts for NAD and NMN in extracts from C2C12 LentiCRISPR whole cells (C) and isolated mitochondria (D) following a 4 hr incubation with isotopically-labeled NR or NAR tracer. (Single measuements). (E–F) Fractional labeling of metabolites (NAD and NMN) measured in C2C12 LentiCRISPR whole cells (E) and isolated mitochondria (F) after a 4 hr incubation with isotopically-labeled NR or NAR tracer. (Single measurements). (G) Fractions of double-labeled NAD and NMN measured in C2C12 LentiCRISPR whole cell and mitochondrial lysates following 4 hr incubation with isotope-labeled NAR (means ± SEM). (N = 3). (H) Immunoblot confirming NMNAT1 knockout in CRISPR C2C12 cell line. (*, p<0.05; **, p<0.001; 2-tailed, unpaired Student’s t-test versus R26).