SUMMARY
Enhancer-derived RNAs are thought to act locally by contributing to their parent enhancer function. Whether large domains of clustered enhancers (super-enhancers) also produce cis-acting RNAs, however, remains unclear. Unlike typical enhancers, super-enhancers form large spans of robustly transcribed chromatin, amassing capped and polyadenylated RNAs that are sufficiently abundant to sustain trans functions. Here, we show that one such RNA, alncRNA-EC7/Bloodlinc, is transcribed from a super-enhancer of the erythroid membrane transporter SLC4A1/BAND3 but diffuses beyond this site. Bloodlinc localizes to trans-chromosomal loci encoding critical regulators and effectors of terminal erythropoiesis and directly binds chromatin- organizing and transcription factors, including the chromatin attachment factor HNRNPU. Inhibiting Bloodlinc or Hnrnpu compromises the terminal erythropoiesis gene program, blocking red cell production, whereas expressing Bloodlinc ectopically stimulates this program and can promote erythroblast proliferation and enucleation in the absence of differentiation stimuli. Thus, Bloodlinc is a transacting super-enhancer RNA that potentiates red blood cell development.
In Brief
Alvarez-Dominguez et al. report that unlike typical enhancers, super-enhancers accumulate RNAs sufficiently abundant and stable to sustain trans functions. One such RNA, Bloodlinc, associates with chromatin and transcription cofactors, binds distal loci encoding key effectors of terminal erythropoiesis, and modulates their expression, thereby potentiating red blood cell development in trans.
INTRODUCTION
Cell development demands precise coordination of cell-type-and state-specific gene expression programs. In eukaryotes, this is achieved by networks of transcription effectors converging at DNA promoter and enhancer regions in response to developmental and environmental signals (Buecker and Wysocka, 2012). By virtue of their engagement with the transcription machinery, active enhancer regions are pervasively transcribed into non-coding RNAs, which in some cases have been shown to contribute locally to enhancer activity (Lam et al., 2014; Natoli and Andrau, 2012). Although their mechanisms are poorly understood, enhancer RNAs are thought to be too unstable and/or too lowly expressed to be able to act in trans.
Large domains of clustered enhancers, called stretch or super-enhancers, have been linked to control of genes important for mammalian cell-type-specific processes (Gaulton et al., 2010; Parker et al., 2013; Whyte et al., 2013). Super-enhancers differ from typical enhancer domains by their higher density of transcription effectors and enhancer-associated histone marks. Not surprisingly, super-enhancers amass higher levels of transcription machinery and are more highly transcribed (Hnisz et al., 2013; Vahedi et al., 2015).
Whether the mechanisms of super-enhancers involve cis-acting RNAs is not understood. Although there is evidence that super-enhancer RNAs can influence target gene activation via modest but detectable effects on chromatin looping and transcription factor recruitment (Sigova et al., 2015; Xiang et al., 2014), it is unclear whether such effects depend on the RNAs themselves or on the act of their transcription. The higher transcriptional activity of super-enhancers relative to typical enhancer domains, on the other hand, suggests that some may produce RNAs that are sufficiently stable and/or abundant to sustain trans functions.
Here, we profile super-enhancers in primary mouse erythroid cells and find that they accumulate capped and polyadenylated RNAs to greater levels than typical enhancers. We focus on one such RNA, previously cataloged as alncRNA-EC7 (Alvarez-Dominguez et al., 2014) or Bloodlinc (Paralkar et al., 2014), which is transcribed from a conserved super-enhancer needed for high-level expression of the erythroid membrane transporter BAND3/SLC4A1. We show that Bloodlinc diffuses beyond its parent super-enhancer domain to multiple trans-chromosomal loci encoding critical regulators and effectors of terminal erythropoiesis that are reciprocally regulated upon Bloodlinc inhibition or ectopic expression. Further, we identify factors involved in chromatin organization and transcription regulation among Bloodlinc’s binding partners, including the chromatin attachment factor HNRNPU, illuminating how Bloodlinc regulates its targets. Importantly, inhibiting Bloodlinc or Hnrnpu impairs the global gene program of terminal erythropoiesis, blocking red cell production, whereas ectopically expressed Bloodlinc stimulates erythroid gene expression and is able to prompt erythroid progenitors to proliferate and terminally differentiate into enucleated reticulocytes in the absence of differentiation stimuli. Thus, we characterize Bloodlinc as a trans-acting super-enhancer RNA capable of potentiating red blood cell development.
RESULTS
Global Characterization of Erythroid Super-Enhancers and Their RNA Products
To catalog mouse erythroid enhancers, we exploited global maps of DNA accessibility assayed by DNase I hypersensitivity (DHS) in E14.5 fetal liver CD117− CD71+ TER119+ erythroblasts (Stamatoyannopoulos et al., 2012). We classified DHS sites on the basis of their overlap with gene promoter-proximal (transcription start site [TSS] ± 2 kb) regions into promoter or putative enhancer sites. As expected, enhancers but not promoters have a high H3K4me1/H3K4me3 ratio and are bidirectionally transcribed, while both active promoters and enhancers enrich for H3K27ac and RNA polymerase II (Figures S1A and S1B). We then linked clustered enhancer sites and ranked the resulting domains by their H3K4me1 signal density to find those with exponentially higher enrichment than the rest, which identifies super-enhancers (Whyte et al., 2013) (Figure 1A; Table S1). We identified 450 super-enhancers, comprising large domains (median size 15.2 kb) that amass ~63% of the total enhancer-associated H3K4me1 signal in red blood cells (Figures S1C and S1D). The genes closest to or overlapping super-enhancers enrich for erythroid-specific proteins critical for red cell maturation and functional specialization, both long known (e.g., α-/β-globins and BAND3/SLC4A1) and more recently characterized (e.g., HIPK2, RBM38, CPEB4) (Figures 1A, S1E, and S1F). Importantly, super-enhancer enhancer constituents enrich for binding of master erythroid transcription factors GATA1 and TAL1 (Figure S1G), and the catalog of super-enhancer-associated genes is highly robust to the particular transcription effector or histone mark used to define super-enhancers (Figures S1H and S1I; Table S1).
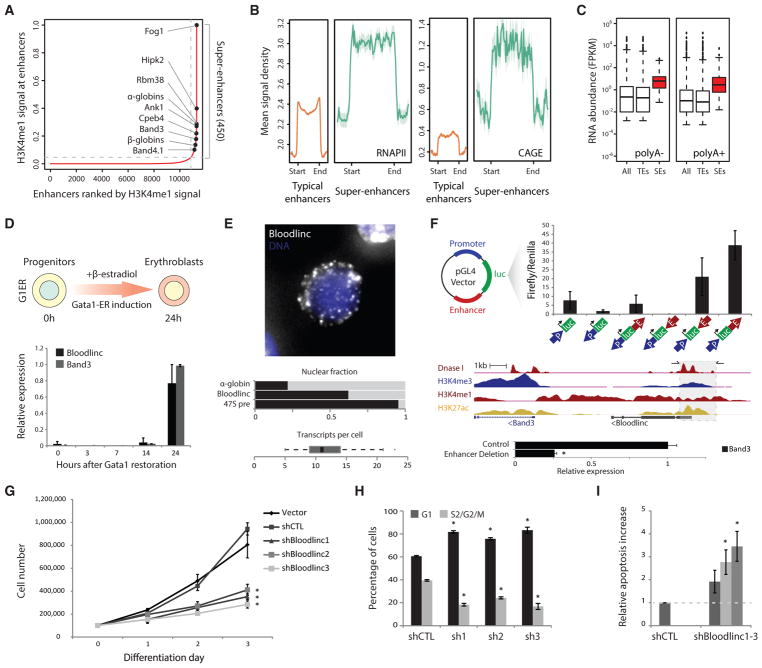
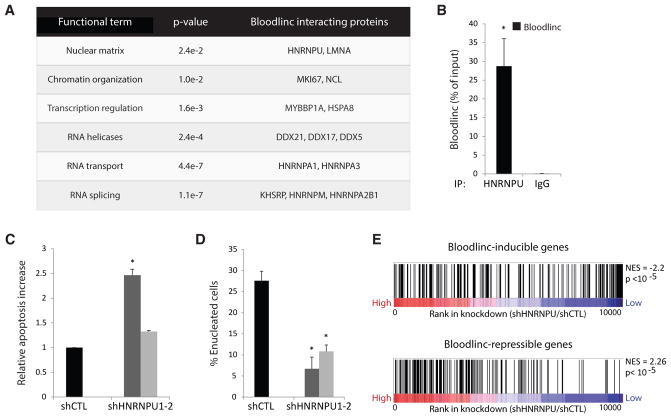
Figure 1. Bloodlinc Is Transcribed from a Band3 Super-Enhancer and Is Required for Survival and Proliferation of Differentiating Erythroblasts.
(A) The distribution of normalized H3K4me1 signal enrichment across erythroid enhancer domains reveals a subset with exponentially higher levels than the rest, comprising 450 super-enhancers (SEs). Erythroid-important genes associated with SEs are highlighted.
(B) Erythroid super-enhancers are more highly transcribed than typical enhancer domains. Metagene plots show mean RNA polymerase II and CAGE signal density across typical- and super-enhancer domains ± 2.5 kb flanking regions.
(C) Erythroid super-enhancers accumulate more RNA than typical enhancer domains. Boxplots show poly(A)− and poly(A)+ RNA abundance (fragments per kilobase of transcript per million mapped reads [FPKM]) for all enhancers, typical enhancers (TEs), and super-enhancers (SEs).
(D) Bloodlinc and Band3 show concordant and late-stage activation upon restoration of GATA1 by β-estradiol treatment of G1ER cells. Data are mean ± 95% confidence interval.
(E) Bloodlinc is a diffuse, pancellular RNA in late-stage erythroblasts. Top: maximum z stack projection of RNA FISH fluorescence microscopy image. Middle: proportion of Bloodlinc transcripts in the nucleus, compared with α-globin and 47S pre-rRNA. Bottom: distribution of per-cell Bloodlinc transcript counts across n = 65 cells with more than zero transcripts.
(F) Bloodlinc’s templating enhancer is necessary and sufficient for high-level Band3 erythroid expression in vivo. Top: enhancer reporter assays in MEL cells evidence orientation-independent enhancer function. Middle: strategy for enhancer deletion (shaded area) using flanking CRISPR/Cas9 single-guide RNAs (sgRNAs), with their location shown by horizontal arrows at the top. Bottom: Band3 high-level induction is abolished in sgRNA-transduced MEL cells. Data are mean ± SEM from n = 3 replicate measurements.
(G) Bloodlinc inhibition blocks proliferation during terminal erythroid differentiation. Shown are live cell numbers in ex vivo-differentiated erythroid cultures transduced with empty vector, non-targeting shRNA, or Bloodlinc-targeting shRNAs. Data are mean ± SEM from n = 3 experiments with n = 1 or 2 replicate measurements each.
(H) Bloodlinc KD cells accumulate in G1 phase after 24 hr of ex vivo differentiation. Data are mean ± SEM from n = 3 experiments.
(I) Elevated apoptosis of Bloodlinc KD cells after 24 hr of ex vivo differentiation. Data are mean ± SEM from n = 3 experiments.
*p < 0.05 relative to control, t test. See also Figures S1–S3 and Table S1.
To profile transcription of erythroid super-enhancers, we probed strand-specific maps of poly(A)+ and poly(A)− RNA and of poly(A)+ RNA 5′ caps from fetal liver TER119+ erythroblasts (Alvarez-Dominguez et al., 2014; Marques et al., 2013). We find that compared with typical enhancers, super-enhancers harbor greater density of RNA polymerase II and produce more poly(A)+ and poly(A)− RNA (p < 10−15 for all, Kolmogorov-Smirnov test) (Figures 1B and 1C). These features are exemplified by the well-known β-globin locus control region, which abundantly produces bidirectional and capped RNAs compared with typical enhancers within the erythroid-important but broadly expressed Mbnl1 gene (Figure S1J).
Bloodlinc Is a Conserved RNA Transcribed from a Band3 Super-Enhancer
To prioritize sufficiently abundant and stable super-enhancer RNAs for functional studies, we quantified RNA levels across all super-enhancer domains and ranked them on the basis of their normalized expression (Table S1). The most abundant extragenic RNA belongs to a super-enhancer encompassing Band3/Slc4a1, which encodes the main red cell membrane anion exchanger. We previously cataloged this RNA as alncRNA-EC7 (Alvarez-Dominguez et al., 2014) (also known as Bloodlinc; Paralkar et al., 2014), a capped and polyadenylated, 3.7 kb spliced long non-coding RNA (lncRNA) bidirectionally transcribed from a 5.2 kb H3K4me1-enriched, open chromatin region ~10 kb upstream of Band3 that is bound by GATA1, TAL1, and KLF1 (Figure 1F, middle, and Figures S2A and S2B). Detecting Bloodlinc by RNA fluorescence in situ hybridization (FISH) in mixed populations of pro-erythroblasts and erythroblasts at early- to mid-stages of differentiation reveals mostly focal sites of nascent transcription in the nucleus (Alvarez-Dominguez et al., 2014). Bloodlinc’s high-level induction occurs only in late-stage differentiating erythroblasts (Figure 1D), however, and detection in these cells reveals it as a diffuse, pancellular RNA (~12 ± 2 transcripts per cell, range 5–48 transcripts; Figure 1E), consistent with previous observations (Paralkar et al., 2014). Importantly, the Bloodlinc transcript is conserved in primary sequence between mouse and human (Figures S2A and S2B) and is required for red cell maturation (Alvarez-Dominguez et al., 2014; Paralkar et al., 2014).
Bloodlinc’s templating DNA physically contacts the Band3 promoter (Figure S2C), and both are coordinately activated by GATA1 in late-stage erythroblasts (Figures 1D and S2D) in a lineage-specific manner (Figure S2F), suggesting that Bloodlinc derives from a super-enhancer that drives Band3 erythroid expression. To test this, we conducted both transient enhancer assays and genetic deletion studies in mouse erythroleukemia (MEL) cells, which robustly induce Bloodlinc upon differentiation (Figure S2E). For enhancer assays, we cloned DNA fragments comprising the GATA1/TAL1/KLF1-bound segments within the Band3 promoter or the Bloodlinc enhancer into luciferase reporter constructs and transfected them into MEL cells subsequently induced to differentiate. The enhancer conveyed ~20- to 40-fold greater reporter expression than the promoter segment alone, independent of its orientation but dependent on concordant orientation between promoter and reporter gene (Figure 1F, top). To delete the enhancer in its native chromatin context, we cloned separate CRISPR/Cas9 guide RNAs flanking the enhancer segment into puromycin-selectable vectors and transduced them into MEL cells cultured under puromycin selection for 7 days prior to induction of differentiation. Assaying Band3 levels in double-transduced cells revealed ~75% expression reduction (Figure 1F, bottom). These findings demonstrate that Bloodlinc’s templating DNA is a bona fide enhancer, as it is both necessary and sufficient to enhance Band3 expression in an orientation-independent manner in erythroid cells. Thus, Bloodlinc is a bona fide enhancer-derived RNA.
Bloodlinc Is Critical for Survival and Proliferation during Terminal Erythropoiesis
To investigate the basis for Bloodlinc’s requirement during terminal erythropoiesis, we depleted it by ~50%–60% by transducing independent short hairpin RNAs (shRNAs) into highly enriched populations of fetal liver erythroid progenitors subsequently induced to terminally differentiate in culture (Figure S3A). Bloodlinc knockdown markedly blocked terminal erythroid proliferation (Figures 1G and S3B; p < 10−3 for all, t test) and caused cells to accumulate in the G1 phase after 24 hr of culture in differentiation medium (Figures 1H and S3C; p < 10−3–10−2, t test). After 48 hr, pulse-labeling with a reactive deoxynucleotide analog (EdU) revealed that more cells had accumulated in S phase (Figure S3D), consistent with a slow division rate and ineffective erythropoiesis. Accordingly, we measured a corresponding ~2- to 4-fold increase in apoptotic or dead cells during the differentiation time course (Figures 1I, S3E, and S3F). To directly assess generation of mature, enucleated reticulocytes, we studied cell DNA content, morphology, and presence of the differentiation surface marker TER119. Inhibiting Bloodlinc severely impaired reticulocyte generation, reducing enucleation efficiency of surviving cells by ~80% (Figure S3G; p < 10−3–10−2, t test), and led to accumulation of larger, nucleated cells, evidencing retention of immature erythroblasts (Figure S3H). TER119 induction was unaffected, consistent with normal initiation of the terminal differentiation program (at the time when Bloodlinc expression is induced). Thus, Bloodlinc is needed for survival and proliferation of differentiating erythroblasts, explaining its requirement for terminal erythropoiesis.
Bloodlinc Is Not Confined to Its Parent Super-Enhancer Domain
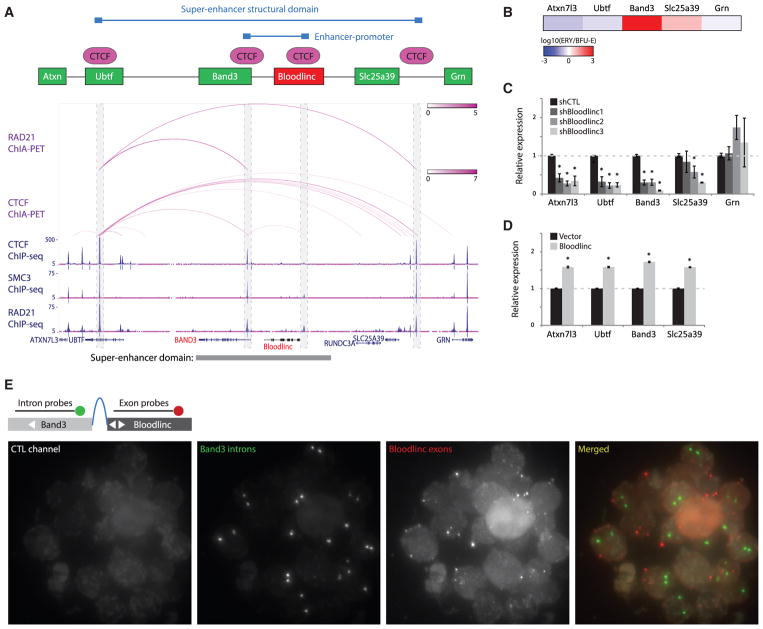
Unlike Bloodlinc, the BAND3 anion exchanger is in fact dispensable for terminal erythropoiesis (Ji and Lodish, 2012; Peters et al., 1996), suggesting that Bloodlinc may not simply act in cis via Band3 activation. To explore this possibility, we initially sought to find additional targets of its templating super-enhancer. Activity of super-enhancers is typically confined to closed chromatin loops formed by interacting sites of the CTCF insulator that are co-bound by cohesin, which limit interactions with chromatin outside the loop to prevent mistargeting nearby genes (Dowen et al., 2014). Bloodlinc’s parent super-enhancer is indeed bounded by CTCF (Figure S2C) and by the cohesin subunits RAD21 and SMC3 in both human and mouse erythroid cells (Figures 2A and S4A), and maps of chromatin interactions involving these factors (Figures 2A and S4A) or of general chromosome conformation in human erythroleukemia cells (Figure S4B) reveal that the domain acquires a closed-loop conformation, with no interactions between internal CTCF- or cohesin-bound sites and external chromatin and no inter-chromosomal interactions (ENCODE Project Consortium, 2012; Heidari et al., 2014; Li et al., 2012). The two other genes within this insulated domain that are active in erythroblasts besides Band3 (Ubtf and Slc25a39) (Figure S4C) thus represent the other possible super-enhancer targets. Unlike Band3, however, these genes lack high-level induction unique to red blood cells (Figures 2B and S4D) and show only weak contacts with the Bloodlinc enhancer (Figure S2C), suggesting that Bloodlinc’s physiological effects cannot be accounted for by the possible targets of its parent super-enhancer.
Figure 2. Bloodlinc Is Not Confined to Its Parent Super-Enhancer Domain.
(A) Bloodlinc’s templating SE is located within an insulated structural domain. Interacting sites of the CTCF insulator co-bound by cohesin delimit the SE domain (model shown on top of tracks). Tracks display RAD21 and CTCF ChIA-PET interactions (purple arcs) and RAD21, SMC3, and CTCF chromatin immunoprecipitation sequencing (ChIP-seq) signal (blue density maps) in K562 cells. Gene models are depicted in blue, and the SE is highlighted at the bottom. Outer boxes highlight the limits of the SE domain, and inner boxes highlight interacting sites between Bloodlinc and the Band3 promoter.
(B) Band3 is the only gene within or flanking the SE domain with high-level induction between BFU-Es and TER119+ erythroblasts (ERY).
(C) Inhibiting Bloodlinc in ex vivo-differentiated erythroid precursors downregulates mRNA levels of genes within and outside the SE domain. Data are mean ± SEM from n = 2 experiments with n = 3 replicate measurements each. *p < 0.05 relative to control, t test.
(D) Overexpressing Bloodlinc in ex vivo-differentiated erythroid precursors upregulates mRNA levels of genes within and outside the SE domain. Data are mean ± SEM from n = 2 experiments with n = 3 replicate measurements each. *p < 0.05 relative to control, t test.
(E) Bloodlinc diffuses beyond its enhancer-promoter DNA loop of origin. Panels show maximum zstack projections of RNA FISH fluorescence microscopy images labeling Bloodlinc exons (red) and Band3 introns (green) in mixed-stage erythroblasts. Control channel measures background fluorescence.
See also Figure S4.
Bloodlinc inhibition leads to dramatic (~70%–90%) reduction in Band3 expression (Alvarez-Dominguez et al., 2014). Strikingly, depleting Bloodlinc or deleting its templating enhancer also downregulates Ubtf (repressed during erythropoiesis) but not Slc25a39 (erythropoiesis induced) (Figures 2C and S5C). Conversely, ectopically overexpressing Bloodlinc by 1.6-fold its physiological levels invariably upregulated each of these genes as well as Atxn7l3 (erythropoiesis repressed and outside the super-enhancer domain) (Figure 2D), suggesting that Bloodlinc acts beyond the confines of its parent super-enhancer. To test this, we designed RNA FISH probes to simultaneously detect Bloodlinc exons and Band3 introns (which mark the location of its closed DNA loop of origin). Unlike RNAs that remain tethered to their transcription site, we find that Bloodlinc diffuses beyond its domain of origin (Figure 2E), evidencing trans-localization.
Bloodlinc Modulates the Core Gene Program of Terminal Erythropoiesis
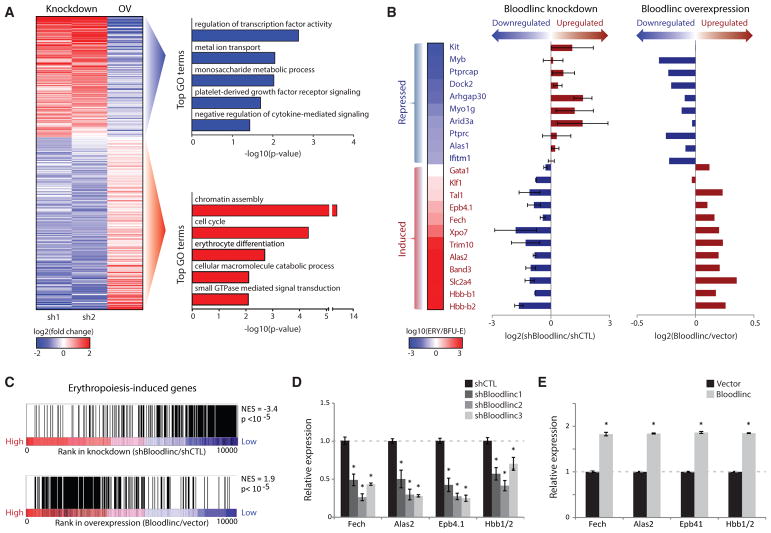
To probe Bloodlinc’s impact on global gene expression during erythropoiesis without bias, we conducted RNA sequencing (RNA-seq) of cultured erythroblasts transduced with shRNAs or Bloodlinc-expressing vector and identified 488 genes that are reciprocally regulated (Table S2). Bloodlinc-repressible genes comprise mostly genes that are normally suppressed with terminal differentiation (e.g., Kit, Myb) (Figures 3A and 3B, top), consistent with its requirement for effective erythropoiesis. Conversely, Bloodlinc-inducible genes enrich for general roles in cell cycling and for specific roles in erythrocyte differentiation (e.g., Gata1/Tal1/Klf1 and β-globin) (Figures 3A and 3B, bottom). Gene set enrichment analysis further revealed general down-and upregulation of erythropoiesis-induced genes upon Bloodlinc knockdown and overexpression, respectively (Figure 3C), which we corroborated by qPCR for select markers in Bloodlinc-depleted cells (Figure 3D), in cells deleted for its templating enhancer (Figure S5D), and in Bloodlinc-overexpressing cells (Figure 3E). Using pathway analysis, we separately identified the GATA1/TAL1/KLF1 regulatory cascade as the most likely mechanism explaining genes downregulated in Bloodlinc-knockdown cells and activated in overexpression cells (p < 10−8–10−5, Fisher’s exact test) (Figure S5A; Table S2). Conversely, genes upregulated upon Bloodlinc knockdown but repressed upon overexpression are most likely explained by modulation of P53 signaling, consistent with Bloodlinc’s requirement for cell survival during terminal erythropoiesis. Accordingly, gene set enrichment analysis indicated global modulation of GATA1/TAL1 and P53 targets upon Bloodlinc modulation (Figure S5B). Thus, Bloodlinc can trans-modulate expression of hundreds of genes important for terminal erythropoiesis, likely reflecting both direct and indirect effects.
Figure 3. Bloodlinc Modulates the Core Gene Program of Terminal Erythropoiesis.
(A) Left: expression change (log2 fold change over control) of 488 genes differentially and reciprocally regulated upon Bloodlinc shRNA-mediated inhibition or ectopic overexpression (OV) after 24 hr of ex vivo differentiation. Right: top non-redundant gene ontology (GO) biological processes enriched among reciprocally regulated genes.
(B) Bloodlinc modulation impacts erythropoiesis-repressed (top) and erythropoiesis-induced genes (bottom). Expression changes for select genes (left) during normal differentiation is shown as the log10 expression ratio between BFU-Es and TER119+ erythroblasts (ERY). Their expression changes upon Bloodlinc depletion or overexpression (right) are shown as the log2 expression ratio between treated versus control cells. Depletion data are pooled from two separate shRNAs.
(C) Erythropoiesis-induced genes are Bloodlinc-inducible. Gene set enrichment analysis of genes induced in erythroblasts versus progenitors (Alvarez-Dominguez et al., 2014) in Bloodlinc-depleted versus Bloodlinc-overexpressing cells. NES, normalized enrichment score.
(D and E) qPCR detection of erythroid markers reciprocally regulated in Bloodlinc-depleted (D) versus Bloodlinc-overexpressing (E) cells. Data are mean ± SEM from n = 2 experiments with n = 3 replicate measurements each. *p < 0.05 relative to control, t test.
See also Figure S5 and Table S2.
Bloodlinc Localizes to trans-Chromosomal Loci Encoding Key Erythropoiesis Modulators
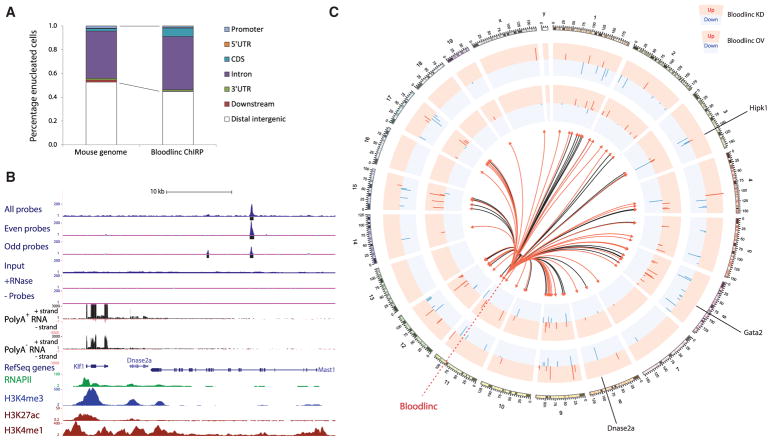
To identify Bloodlinc’s direct targets, we conducted chromatin isolation by RNA purification followed by sequencing (ChIRP-seq) (Chu et al., 2011) in MEL cells, using a biotin-labeled version of the same probes used for RNA FISH that robustly and specifically retrieve Bloodlinc. As controls, we repeated ChIRP-seq after splitting the probes into two separate pools and compared results with RNase-treated and no probe conditions (Figures S6A and S6B). We detected 269 high-confidence Bloodlinc occupancy sites genome-wide (Table S3; Supplemental Experimental Procedures), which are overrepresented in genic regions (p < 10−14, Fisher’s exact test; Figure 4A). The genes mapping to or nearest to these regions include key erythroid modulators, comprising proteins involved in RAS signaling, which regulates erythropoiesis (Zhang et al., 2003), transcription factors that direct terminal differentiation, such as KLF1, and direct effectors of erythrocyte function, such as SLC11A2 (an essential iron transporter). Bloodlinc localization is focal (median binding peak size 240 bp), reminiscent of transcription factor binding, and can occur within regulatory regions distal to erythroid-active genes, as with the Klf1/Dnase2a locus (Figure 4B), or proximal to them, as with the Gata2 locus (Figure S6E). Motif analysis of Bloodlinc binding sites revealed no significant resemblance to known recognition sites of DNA-binding proteins, and we found no significant interaction with (Figure S6C and see below) or overlap with the binding sites of core erythroid transcription factors GATA1, TAL1, or KLF1 (Figure S6D). Importantly, we identified 81 direct gene targets across multiple chromosomes that are reciprocally regulated upon Bloodlinc knockdown and overexpression (Figure 4C; Table S3). These comprised 60 Bloodlinc-induced genes that include Hipk1, a transcription cofactor that modulates erythropoiesis (Hattangadi et al., 2010), and 21 Bloodlinc-repressed genes, including the Tspan2 developmental signaling regulator. These data thus demonstrate that Bloodlinc binds to and regulates trans-chromosomal loci encoding key terminal erythropoiesis modulators.
Figure 4. Bloodlinc Localizes to trans-Chromosomal Loci Encoding Key Erythropoiesis Modulators.
(A) Bloodlinc genomic occupancy is enriched in genic regions.
(B) Bloodlinc occupancy is focal, specific, and can occur within regulatory regions distal to erythroid-active genes, such as the Klf1/Dnase2a locus. Top tracks display signal density for Bloodlinc ChIRP-seq with all probes or with split probe pools, as well as input, RNase-treated, and no probe controls. Bottom tracks display strand-specific maps of poly(A)+ RNA and poly(A)− RNA signal density, and occupancy maps of RNA polymerase II, H3K4me3, H3K4me1, and H3K27ac. Gene models are depicted in blue.
(C) Bloodlinc diffuses from its transcription site to trans-chromosomal gene targets that are reciprocally regulated upon Bloodlinc knockdown and overexpression. Tracks show expression changes (log2 fold change over control) of 81 genes differentially and reciprocally regulated upon Bloodlinc shRNA-mediated knockdown (KD; outer tracks) or ectopic overexpression (OV; inner tracks), inscribed at their respective genomic locales. Depletion data are pooled from two separate shRNAs. The center of the circle depicts Bloodlinc diffusion to its ChIRP-seq occupancy sites as directional links from its genomic locus. Links to occupancy sites intersecting genic regions are highlighted in red.
See also Figure S6 and Table S3.
Bloodlinc Interacts with Chromatin-Organizing and Transcription Factors
To investigate how Bloodlinc modulates its targets, we conducted comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) (Chu et al., 2015) in MEL cells. As controls, we also performed ChIRP-MS of Dleu2, another erythroid differentiation-induced lncRNA residing in the nucleus (Alvarez-Dominguez et al., 2014), and compared results before and after high-level Bloodlinc and Dleu2 induction. Using stringent criteria (Supplemental Experimental Procedures), we identified 33 high-confidence Bloodlinc-binding proteins (Figure 5A; see full list of enriched proteins with peptide counts in Table S4). These comprised Bloodlinc-specific as well as general RNA processing proteins (involved in RNA unwinding, splicing, packaging and transport) that non-specifically associate with Bloodlinc, Dleu2, and other RNAs (Neat1/Malat1/U1/U2/Xist, assayed in different cells by the same method; Table S4). The Bloodlinc-specific interactors are enriched for roles in chromatin organization (e.g., MKI67, LMNA) and transcriptional regulation (e.g., MYBBP1A, HSPA8), consistent with Bloodlinc’s ability to bind chromatin and modulate gene expression. The MYBBP1A protein binds the essential hematopoietic regulator MYB and can act as a transcriptional coactivator or corepressor, the latter by recruiting the SIN3-HDAC1 histone deacetylase complex (Hara et al., 2009; Jones et al., 2002), while HSPA8 has been implicated in transcriptional silencing by suppressing coactivator recruitment (Yahata et al., 2000). MKI67 and LMNA control gene expression at the chromatin level, on the other hand: MKI67 directs heterochromatin organization in proliferating cells (Sobecki et al., 2016), and LMA, a major nuclear lamina component, is needed for heterochromatin formation (Burke and Stewart, 2013) and for terminal erythropoiesis (Li et al., 2016).
Figure 5. Bloodlinc Interacts with Chromatin-Organizing and Transcription Factors.
(A) Bloodlinc high-confidence interactors enrich for specific functional classes (p < 0.05, Fisher’s exact test); select members of each class are listed to the right (see Table S4 for full list of interactors with peptide counts).
(B) Bloodlinc specifically and strongly interacts with HNRNPU. Bloodlinc retrieval was assayed by qPCR in immunoprecipitates of endogenous HNRNPU from MEL cells and compared with IgG control. Data are mean ± SEM from n = 5 experiments with n = 3 replicate measurements each.
(C) Elevated apoptosis of Hnrnpu KD cells after 24 hr of ex vivo differentiation. Data are mean ± SEM from n = 2 experiments.
(D) Hnrnpu knockdown impairs red cell enucleation. Data are mean ± SEM from n = 3 experiments.
(E) Hnrnpu-depleted cells phenocopy the gene expression changes of Bloodlinc-depleted cells. Gene set enrichment analysis of Bloodlinc-inducible and Bloodlinc-repressible genes in Hnrnpu KD cells. NES, normalized enrichment score.
*p < 0.05 relative to control, t test. See also Figure S6 and Table S4.
Our identification of the nuclear matrix factor HNRNPU was of particular interest because it is known to stabilize RNA-chromatin associations throughout the genome (Hacisuleyman et al., 2014; Hall et al., 2014; Hasegawa et al., 2010). To confirm the Bloodlinc-HNRNPU interaction, we verified that immunoprecipitates of endogenous HNRNPU specifically and strongly enrich for Bloodlinc (~30% of input RNA) (Figure 5B). We then transduced Hnrnpu-targeting shRNAs into primary erythroid progenitors subsequently induced to differentiate in culture and found that, analogous to Bloodlinc inhibition, Hnrnpu knockdown causes elevated apoptosis (Figures 5C and S6F) and blocks reticulocyte generation (Figures 5D and S6G). We thus conducted RNA-seq of Hnrnpu-knockdown cells to evaluate the extent to which their gene expression changes phenocopy those of Bloodlinc-depleted cells and found significant overlap with Bloodlinc-inducible and Bloodlinc-repressible genes (both empirical p < 10−5; Figure 5E). These data suggest that Bloodlinc and HNRNPU are part of a functional ribonucleoprotein complex regulating a common set of genomic targets that is essential for terminal erythropoiesis. Together, our findings support a model whereby Bloodlinc forms stable trans-chromosomal interactions to direct erythroid gene expression by recruiting cohorts of repressive or activating chromatin and transcription cofactors.
Bloodlinc Can Promote Terminal Red Cell Development
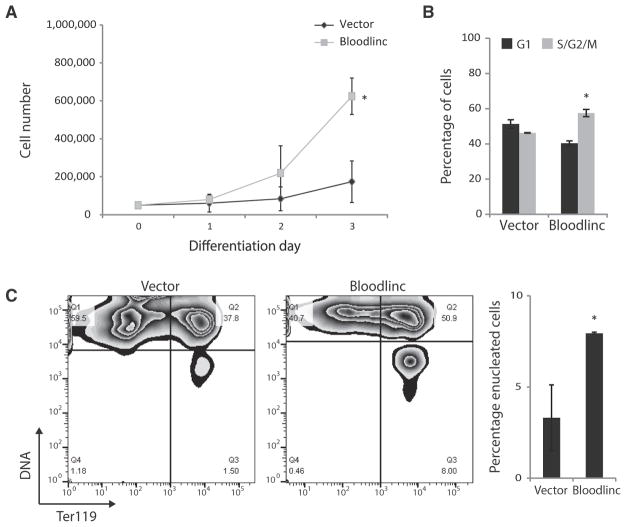
Bloodlinc’s ability to bind to and potentiate expression of genes mediating red cell maturation suggests a capacity to stimulate this developmental process. To test this, we transduced primary TER119-negative erythroid cell cultures, enriched (>90%) for colony-forming unit progenitors and pro-erythroblasts (Alvarez-Dominguez et al., 2017; Zhang et al., 2003), with Bloodlinc-expressing vectors, and cultured them in maintenance medium containing erythropoietin (EPO; to prevent their apoptosis) and stem cell factor (see Experimental Procedures), which does not stimulate their terminal proliferation or differentiation (Hu et al., 2011). Remarkably, ectopically expressed Bloodlinc induces progenitors to undergo substantial proliferation and terminal differentiation, causing a dramatic ~4-fold increase in live cell expansion (Figure 6A) and prompting more to accumulate in the S and G2/M phases of the cell cycle (Figures 6B and S6H), which results in a ~2-fold increase in TER119 induction and enucleated reticulocyte generation (Figure 6C). Furthermore, we find that ectopically expressed Bloodlinc confers some protection to erythroid progenitors against apoptosis caused by culture in maintenance medium lacking EPO (Figure S6I). Thus, Bloodlinc is capable of promoting terminal erythropoiesis in the absence of differentiation stimuli.
Figure 6. Bloodlinc Can Promote Terminal Red Cell Development.
(A) Ectopically expressed Bloodlinc promotes proliferation of erythroid precursors. Shown are live cell numbers of primary erythroid progenitors transduced with empty or Bloodlinc-expressing vector and kept in maintenance medium.
(B) Bloodlinc ectopic expression stimulates erythroid precursor cell cycling. Cells in G1 or in S/G2/M phases after culture for 48 hr in maintenance medium are quantified.
(C) Ectopically expressed Bloodlinc prompts erythroid precursors to terminally differentiate and undergo enucleation. Flow cytometry plots show levels of the differentiation marker TER119 versus DNA content for live primary erythroid progenitors cultured for 48 hr in maintenance medium. Lower right quadrants identify enucleated reticulocytes, quantified to the right.
Data are mean ± SEM from n = 2 experiments with n = 1 or 2 replicate measurements each. *p < 0.05 relative to control, t test. See also Figure S6.
DISCUSSION
Determining whether enhancer RNAs are inert byproducts of enhancer activity or functional players in cell physiology has generated much interest. Recent evidence suggests that some enhancer RNAs can foster activation of nearby genes (Lam et al., 2013; Li et al., 2013; Melo et al., 2013). Depleting enhancer RNAs, however, does not seem to affect transcription factor recruitment, RNA polymerase II loading, or histone marking at enhancers (Hah et al., 2013; Johnson et al., 2003; Mousavi et al., 2013), suggesting that RNA is not needed for enhancer formation. Rather, roles for enhancer RNAs appear limited to stabilizing local chromatin loops, accessibility, and transcription factor binding (Li et al., 2013; Sigova et al., 2015).
Whether similar principles apply to RNAs emanating from large regions of clustered enhancers (super-enhancers) is of particular interest. Chromatin accessibility and transcription factor binding stabilization are particularly important at super-enhancers, as they are densely occupied by transcriptional effectors and are highly sensitive to their perturbation (Hnisz et al., 2013; Lovén et al., 2013; Whyte et al., 2013). On the other hand, the presence of extended open-chromatin spans that are highly transcribed at super-enhancers may yield RNAs with sufficient abundance/stability to sustain broad functions in trans. Systematic dissection of super-enhancer RNA cellular functions has been lacking, however, rendering their contributions to cell development and physiology poorly understood.
Here, we characterize super-enhancers and their RNA products in erythroid cells and focus on the most abundant extragenic one to investigate its cellular functions. We had previously cataloged this RNA as alncRNA-EC7/Bloodlinc, which is transcribed from a Band3 super-enhancer and is essential for red cell maturation, and hypothesized that it could act in part by mediating Band3 induction in cis. This conclusion was consistent with its requirement for Band3 induction, with their tissue and developmental co-expression, and with the detection of nascent RNA foci in heterogeneous populations of pro-erythroblasts and early-to mid-stage differentiating erythroblasts (Alvarez-Dominguez et al., 2014). However, high-level Bloodlinc induction is exquisitely specific to late-stage differentiating erythroblasts, and detecting Bloodlinc in these cells reveals a diffuse, pancellular distribution (this study and Paralkar et al., 2014). We estimate that Bloodlinc exists at ~5–50 copies per late-stage erythroblast, which is ~10-fold greater than the median lncRNA level in any tissue (Derrien et al., 2012). This level is intermediate between that of HOTTIP (less than one copy per cell) (Wang et al., 2011), which interacts with and modulates genes only in very close proximity, and that of XIST (~50–100 copies per cell) (Sunwoo et al., 2015), which interacts with and silences ~1,000 genes, thus suggesting an intermediate capacity for Bloodlinc to locate and regulate hundreds of genes. Indeed, we show that Bloodlinc interacts with >200 loci encoding critical regulators and effectors of terminal erythropoiesis, including but not limited to Band3, which are reciprocally regulated upon Bloodlinc inhibition or ectopic expression. Moreover, we find that ectopically expressed Bloodlinc is capable of prompting erythroid progenitors to proliferate and terminally differentiate into enucleated reticulocytes in the absence of differentiation stimuli. These findings thus demonstrate that Bloodlinc has broad physiological roles by acting in trans to modulate a developmentally important gene program.
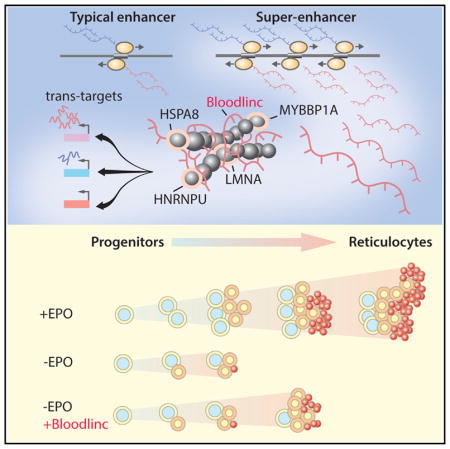
Our report of a trans-acting super-enhancer RNA offers a distinct mechanistic paradigm for enhancer RNA function. Unlike enhancer RNAs that act by stabilizing chromatin contacts or transcription factor binding at their parent enhancer-promoter loops (Lai et al., 2013; Li et al., 2013), Bloodlinc does not seem to interact with the mediator or cohesin complexes (not shown) or to trap the transcription factor that binds its templating super-enhancer. Instead, using an unbiased method for the comprehensive identification of Bloodlinc-binding proteins by mass spectrometry (ChIRP-MS), we identified nuclear matrix proteins and a group of chromatin-organizing and transcription cofactors as Bloodlinc’s specific binding partners. We verified a strong interaction between Bloodlinc and the matrix attachment factor HNRNPU and found that inhibiting HNRNPU phenocopies the gene expression and physiological effects of Bloodlinc inhibition, implying a functional link. HNRNPU fulfills broader roles in nuclear organization, however, such that the effects of its knockdown must reflect perturbation of many functions in addition to those exerted in partnership with Bloodlinc.
Taken together, our findings paint a stepwise model for Bloodlinc’s action mechanism. Specifically, Bloodlinc likely accesses its trans genomic targets via chromatin interactions stabilized by HNRNPU and recruits specific combinations of cofactors to stimulate or repress target gene expression. Bloodlinc may stimulate gene expression by scaffolding both chromatin-opening proteins (e.g., DHX9) and transcriptional co-activators (e.g., MYBBP1A), while gene repression likely involves recruitment of transcriptional repressors (e.g., HSPA8) and of proteins that form a stable closed-chromatin state (e.g., MKI67). Our data thus begin to illuminate how a red-cell specific super-enhancer RNA directs a red-cell specific gene program.
How many more super-enhancer RNAs may act in trans? There are hundreds of RNAs that, like Bloodlinc, emerge from highly transcribed super-enhancers and thus accumulate to substantial levels. These include RNAs from the β-globin locus control region that overlap β-globin and have been implicated in its trans-activation (Ashe et al., 1997). It is thus tempting to speculate that the prevalence of large and highly transcribed open-chromatin regions among super-enhancers provides a general platform where new RNA genes with broad cellular functions can evolve.
EXPERIMENTAL PROCEDURES
Oligos and published data sets used in this study are listed in Tables S5 and S6, respectively.
Cell Isolation, Culture, and Differentiation
All animal experiments were performed with the approval of the Massachusetts Institute of Technology Committee on Animal Care. Mouse fetal liver erythroid cell purification, culture, and differentiation were described previously (Hattangadi et al., 2010; Zhang et al., 2003). The maintenance medium used for expansion of erythroid progenitors without induction of differentiation was StemSpan SFEM II (StemCell Technologies) containing 100 ng/mL recombinant mouse SCF (Amgen), 40 ng/mL recombinant human IGF-1 (R&D Systems), 100 nM dexamethasone, and 2 U/ml EPO (Amgen). The medium used to induce differentiation of erythroid progenitors was Iscove modified Dulbecco’s medium containing 25% FBS (StemCell Technologies), 500 μg/mL holo-transferrin (Sigma-Aldrich), 2 mM L-glutamine, 10 μg/mL recombinant human insulin (Sigma-Aldrich), and 10 U/mL EPO. MEL cell culture and DMSO-induced differentiation are described in the mouse ENCODE Weissman lab protocol (Stamatoyannopoulos et al., 2012).
RNA FISH
RNA FISH and fluorescence microscopy were described previously (Alvarez-Dominguez et al., 2014). Probes were hybridized at 2 ng/μL final concentration (see Supplemental Experimental Procedures for analysis details).
Retroviral Transduction
Isolated erythroid precursors were transduced by murine stem cell virus-based retroviruses as described previously (Hattangadi et al., 2010).
Flow Cytometry Assays
Flow cytometry and cell sorting were conducted on LSR II and Aria FACS machines (BD Biosciences), respectively. Immunostaining and enucleation assays were described previously (Ji et al., 2008; Hattangadi et al., 2010). Cell cycle and apoptosis were analyzed using the Click-iT EdU Pacific Blue Flow Cytometry (Life Technologies) and Annexin V Pacific Blue conjugate (Life Technologies) kits, respectively. Assays were performed on transduced (GFP+) cells, selected by gating (live-cell experiments) or by prior sorting (fixed-cell experiments).
RNA-Seq
Total RNA from erythroid cells transduced with shRNAs, Bloodlinc-expressing, or control vectors was collected using a QIAGEN kit after 24 hr of ex vivo differentiation culture. Strand-specific sequencing libraries were prepared as described (Alvarez-Dominguez et al., 2014) and sequenced on an Illumina HiSeq2000 platform for 40 cycles (see Supplemental Experimental Procedures for analysis details).
ChIRP-Seq and ChIRP-MS
Probes used for RNA FISH were biotin-labeled and used for ChIRP-seq and ChIRP-MS as described previously (Chu et al., 2011, 2015) (see Supplemental Experimental Procedures for analysis details).
Highlights.
Super-enhancers amass capped and polyA+ RNA abundant enough to hold trans functions
Bloodlinc is an SE RNA that diffuses to trans loci encoding key erythroid proteins
Bloodlinc binds chromatin-organizing and transcription factors, including HNRNPU
Bloodlinc can tune gene expression to stimulate red cell production in trans
Acknowledgments
We thank Drs. Wenqian Hu, Jiahai Shi, William Hesse, and Julie Donaghey for reagents and assistance with experiments; Dr. M. Inmaculada Barrasa for bioinformatics support; Tom DiCesare for help with illustrations; and the flow cytometry, mass spectrometry, and genome technology cores at the Whitehead Institute for technical support. J.R.A-D. is a Fellow of the Life Sciences Research Foundation. This research was supported by fellowship Kn1106/1-1 from Deutsche Forschungsgemeinschaft (M.K.) and National Institute of Health grants DK068348 and 5P01 HL066105 (H.F.L.).
Footnotes
ACCESSION NUMBERS
The accession number for the RNA-seq and ChIRP-seq data reported in this paper is GEO: GSE97121.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.05.082.
AUTHOR CONTRIBUTIONS
J.R.A.-D., M.K, and A.A.G. performed experiments. J.R.A.-D., M.K., and H.F.L. designed the research, interpreted results, and wrote the manuscript.
References
- Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123:570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez JR, Zhang X, Hu W. Widespread and dynamic translational control of red blood cell development. Blood. 2017;129:619–629. doi: 10.1182/blood-2016-09-741835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012;28:276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol. 2013;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, Young RA. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, Fackelmayer FO, Lawrence JB. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156:907–919. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Onishi Y, Oishi K, Miyazaki K, Fukamizu A, Ishida N. Molecular characterization of Mybbp1a as a co-repressor on the Period2 promoter. Nucleic Acids Res. 2009;37:1115–1126. doi: 10.1093/nar/gkn1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Hattangadi SM, Burke KA, Lodish HF. Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood. 2010;115:4853–4861. doi: 10.1182/blood-2009-07-235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, Zhang MQ, Snyder MP. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Lodish HF. Ankyrin and band 3 differentially affect expression of membrane glycoproteins but are not required for erythroblast enucleation. Biochem Biophys Res Commun. 2012;417:1188–1192. doi: 10.1016/j.bbrc.2011.12.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol. 2008;10:314–321. doi: 10.1038/ncb1693. [DOI] [PubMed] [Google Scholar]
- Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol Cell Biol. 2003;23:6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LC, Okino ST, Gonda TJ, Whitlock JP., Jr Myb-binding protein 1a augments AhR-dependent gene expression. J Biol Chem. 2002;277:22515–22519. doi: 10.1074/jbc.M200740200. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Shi J, Huang NJ, Pishesha N, Natarajan A, Eng JC, Lodish HF. Efficient CRISPR-Cas9 mediated gene disruption in primary erythroid progenitor cells. Haematologica. 2016;101:e216–e219. doi: 10.3324/haematol.2015.135723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol. 2013;14:R131. doi: 10.1186/gb-2013-14-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Léveillé N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, Dunagin M, Pimkin M, Gore M, Sun D, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123:1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Black BL, et al. NISC Comparative Sequencing Program; National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors; NISC Comparative Sequencing Program Authors. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, Shivdasani RA, Liu SC, Hanspal M, John KM, Gonzalez JM, Brugnara C, Gwynn B, Mohandas N, Alper SL, et al. Anion exchanger 1 (band 3) is required to prevent erythrocytemembrane surface loss but not to form the membrane skeleton. Cell. 1996;86:917–927. doi: 10.1016/s0092-8674(00)80167-1. [DOI] [PubMed] [Google Scholar]
- Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobecki M, Mrouj K, Camasses A, Parisis N, Nicolas E, Llères D, Gerbe F, Prieto S, Krasinska L, David A, et al. The cell proliferation antigen Ki-67 organises heterochromatin. eLife. 2016;5:e13722. doi: 10.7554/eLife.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. Mouse ENCODE Consortium. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Wu JY, Lee JT. The Xist RNA-PRC2 complex at 20- nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc Natl Acad Sci U S A. 2015;112:E4216–E4225. doi: 10.1073/pnas.1503690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SC, Erdos MR, Davis SR, Roychoudhuri R, Restifo NP, Gadina M, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata T, de Caestecker MP, Lechleider RJ, Andriole S, Roberts AB, Isselbacher KJ, Shioda T. The MSG1 non-DNA-binding transactivator binds to the p300/CBP coactivators, enhancing their functional link to the Smad transcription factors. J Biol Chem. 2000;275:8825–8834. doi: 10.1074/jbc.275.12.8825. [DOI] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]