Figure 4. Bloodlinc Localizes to trans-Chromosomal Loci Encoding Key Erythropoiesis Modulators.
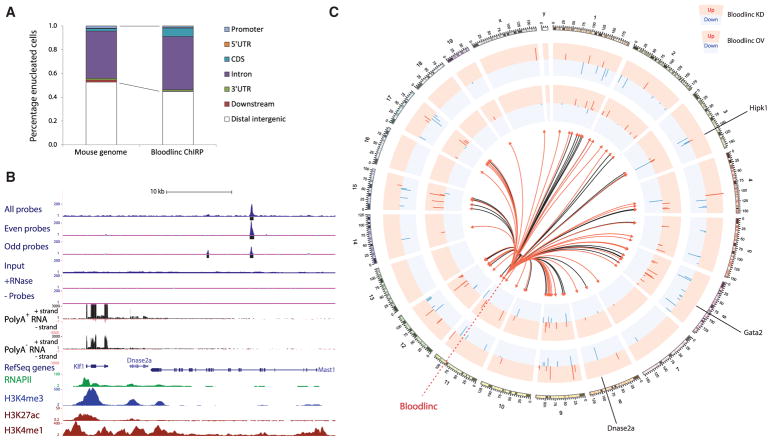
(A) Bloodlinc genomic occupancy is enriched in genic regions.
(B) Bloodlinc occupancy is focal, specific, and can occur within regulatory regions distal to erythroid-active genes, such as the Klf1/Dnase2a locus. Top tracks display signal density for Bloodlinc ChIRP-seq with all probes or with split probe pools, as well as input, RNase-treated, and no probe controls. Bottom tracks display strand-specific maps of poly(A)+ RNA and poly(A)− RNA signal density, and occupancy maps of RNA polymerase II, H3K4me3, H3K4me1, and H3K27ac. Gene models are depicted in blue.
(C) Bloodlinc diffuses from its transcription site to trans-chromosomal gene targets that are reciprocally regulated upon Bloodlinc knockdown and overexpression. Tracks show expression changes (log2 fold change over control) of 81 genes differentially and reciprocally regulated upon Bloodlinc shRNA-mediated knockdown (KD; outer tracks) or ectopic overexpression (OV; inner tracks), inscribed at their respective genomic locales. Depletion data are pooled from two separate shRNAs. The center of the circle depicts Bloodlinc diffusion to its ChIRP-seq occupancy sites as directional links from its genomic locus. Links to occupancy sites intersecting genic regions are highlighted in red.
See also Figure S6 and Table S3.