Figure 3.
The Pattern of Cell Activity Depended on Experimental Conditions
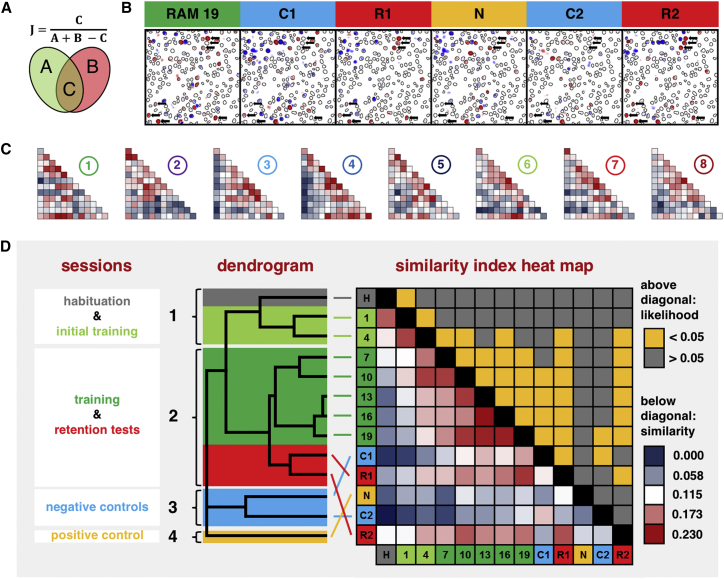
(A) The similarity index J was calculated as the ratio of elements in common C and the sum of all available unique elements A + B − C.
(B) A representative example of a population of cells tracked throughout the study. The outlines denote detected cells, while the coloring denotes the signal difference between timepoint 3 and timepoint 1, with red denoting an increase and blue denoting a decrease in fluorescence. The arrows point to putative “engram” cells.
(C) Similarity index heat maps generated for individual animals. The arrangement of sessions is the same as in (D). Each square of the heat map displays the similarity index between two experimental sessions. Warm colors highlight high similarity index values, and cool colors highlight low similarity index values.
(D) Mean similarity index heat map and corresponding dendrogram. The squares below the diagonal of the heat map represent mean similarity indices, while values above the diagonal represent corresponding (log10) p values with those below (Bonferroni-adjusted) α = 0.05 highlighted in orange. The dendrogram to the left reveals the clustering of conditions based on the mean similarity index values. Four clusters were detected: habituation (H) and days 1 and 4 of RAM training in light green; RAM training days 7–19 with the two retention sessions R1 and R2 in dark green; negative control sessions (C1, C2) in blue; and novelty on its own (N) in orange.