Figure 2.
Nuclear Division Cycles during the Coenocytic Growth Are Periodic, and Their Duration Is Independent of Nutrient Concentration
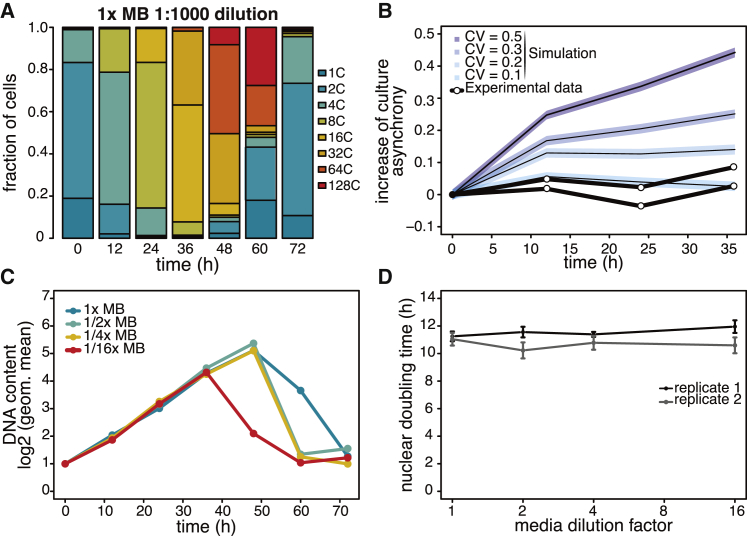
(A) Quantification of DNA content profiles for time course of culture grown in 1× MB at 1:1000 initial dilution of a saturated culture. Approximately 5,000 cells were measured in each population.
(B) Increase of culture asynchrony over time. Asynchrony was calculated as geometric standard deviation of DNA content. Lines with shaded regions represent results from simulation for populations of various cell-to-cell variability in nuclear doubling times. Shaded regions represent the standard deviation from 100 simulations. Black lines with circles represent experimental data for two biological replicates for cultures grown in 1× MB.
(C) Quantification of mean DNA content per time point (expressed as log2 of geometric mean) for cells grown in different media dilutions as indicated.
(D) Nuclear doubling time, calculated by linear regression of mean nuclear content at time points from 0 hr to 36 hr for two sets of biological replicates. Error bars represent standard error of the slope.
See also Figure S3.