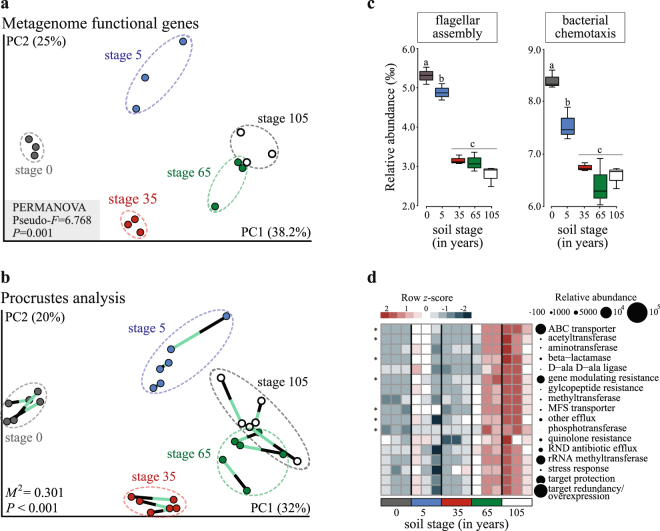
Figure 2.
Functional metagenomes correlate with taxonomic composition across the stages of soil formation and the ‘dispersal’ and ‘competition’ response modus. (a) Principal coordinate analysis (PCoA) based on Bray-Curtis distances calculated from normalized KO annotations. (b) Procrustes analysis depicts a significant correlation between functional metagenomes (Bray-Curtis) and bacterial community composition (Bray-Curtis) across the soil stages. (c) Box plots display the abundance distribution of flagellar assembly (upper) and chemotaxis (down) mechanisms (based on KO annotations) of microbiomes across the soil stages (asterisks denote independent Mann-Whitney U test results performed only for pairwise comparisons between consecutive soil stages, *P < 0.05, **P < 0.001). (d) Heatmap displays the relative abundance (row z-scores) of ARGs by mechanism. ABC, ATP-binding cassette; MFS, major facilitator superfamily; RND, resistance-nodulation-cell division. Circles are proportional to the relative abundance of each ARG mechanism in all samples. Asterisks denote ARG mechanisms that differentially segregated across the soil stages, identified by random forest analysis with Boruta feature selection (average z-scores of 1000 runs >4) (Supplementary Table S5).