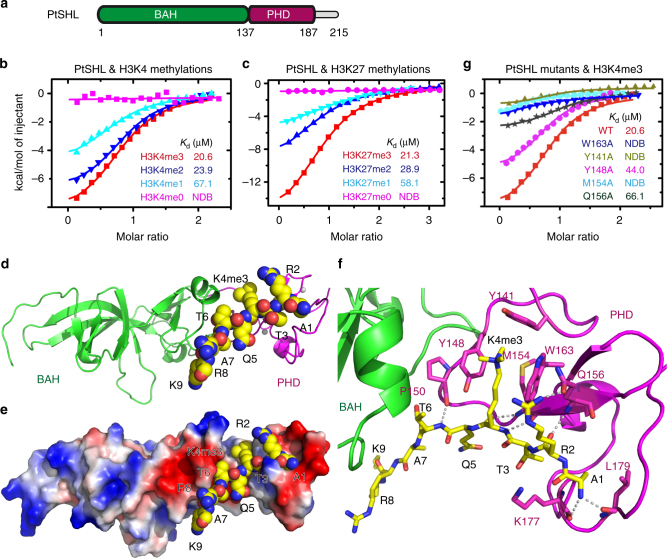
Fig. 2.
Structure of PtSHL in complex with H3K4me3. a The domain architecture of Populus trichocarpa SHL (PtSHL). BAH, bromo-adjacent homology; PHD, plant homeodomain. b, c ITC binding curves for the complex formation between PtSHL and different H3K4 (b) and H3K27 (c) methylated peptides. d Overall structure of PtSHL-H3K4me3 complex with the BAH and PHD domains colored in green and magenta, respectively. The peptides and zinc ions are shown as spacing fill model and silver balls, respectively. e Electrostatic surface view of the PtSHL in complex with H3K4me3. f The detailed interactions between PtSHL and H3K4me3 with the peptide shown in stick model. The hydrogen bonds are highlighted by dashed lines. g ITC binding curves show that the mutations of key residues critical for H3K4me3 binding significantly decrease the binding affinity to the H3K4me3 peptide. NDB, no detectable binding