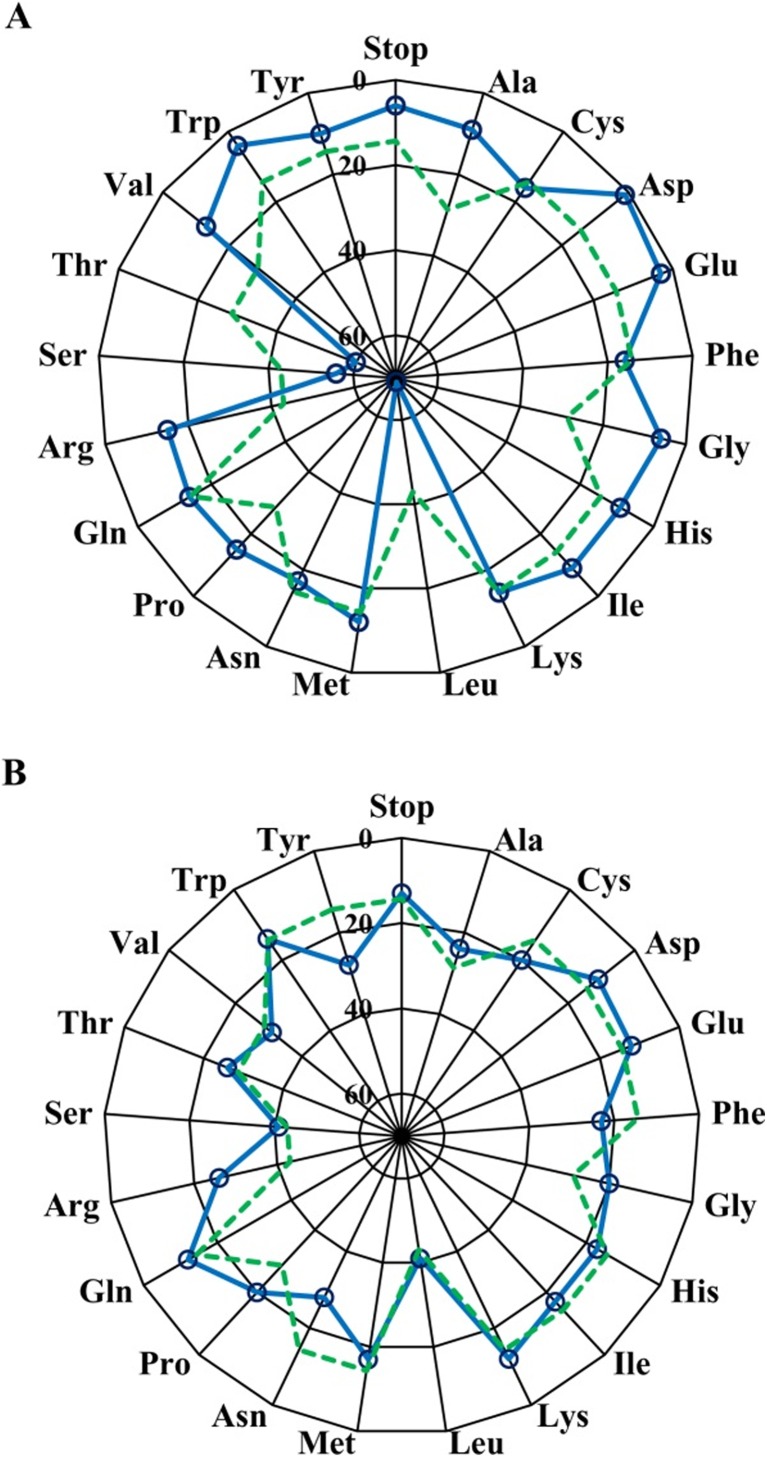
Fig. 5.
Residue distributions across all NNK-based SSM libraries for both one-step PCR technique (a) and two-step PCR approach (b). The aggregate number of occurrences of each amino acid in all five of the saturation libraries is listed with a maximum of 70 in the center of the plot. This scale was chosen based on the number of occurrences of the most commonly observed amino acid (Leu) in the libraries constructed by the one-step PCR approach. The expected aggregate occurrence of each amino acid based on perfectly random replacements at each position is also shown (green dashed lines)