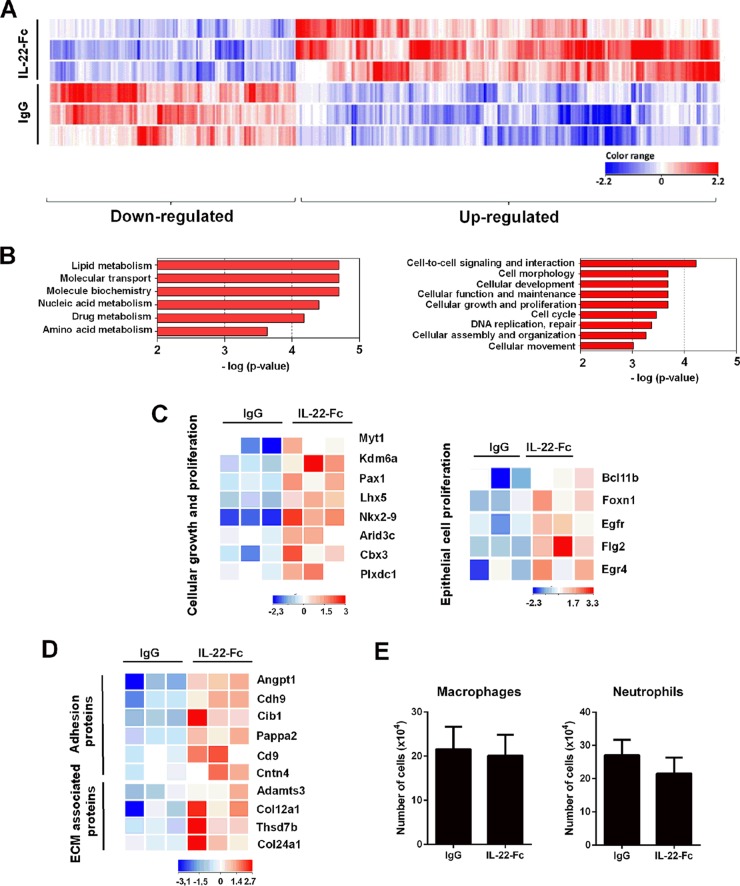
FIG 2.
Transcriptomic signature of IL-22 in the lungs of naive mice. (A) Total RNAs from Ig-treated mice or IL-22-Fc-treated mice (5 μg/animal) were extracted from the lungs at 8 h poststimulation. Hierarchical clustering diagrams showing individual replicates are represented. Normalized intensity values (IL-22-Fc condition compared to IgG condition) are depicted in blue (downregulated) or red (upregulated) (P < 0.05 and fold change of >2). The magnitude of the regulation is illustrated by the intensity of the color. Hierarchical clustering representation indicates a minimal effect of the isotype control in the lung tissue (data not shown). (B) Gene ontological analysis. Significantly enriched pathways are represented as a bar plot (IPA). For panels A and B, data from three mice are shown. (C) Heat maps of genes that were differentially regulated by IL-22 treatment and that belong to the cellular growth and proliferation family (GO:0008283) and, within this family, belonging to the epithelial growth and proliferation subfamily (GO:0050673). Representative genes are noted on the right. Each column represents data from one individual mouse, with three mice per group. (D) Heat maps of genes represented in the families of adhesion proteins (GO:0022610) and extracellular-matrix (ECM)-associated proteins (GO:0031012), both belonging to the morphology family. (E) Naive mice were treated with IL-22-Fc or IgG (5 μg/animal), and 8 h later, cells from whole lungs were analyzed by flow cytometry. The mean number ± standard deviation of alveolar macrophages (CD45+ Siglec F+ CD11blow) and neutrophils (CD11b+ Ly6G+ Siglec F−) are depicted (n = 8; two independent experiments).