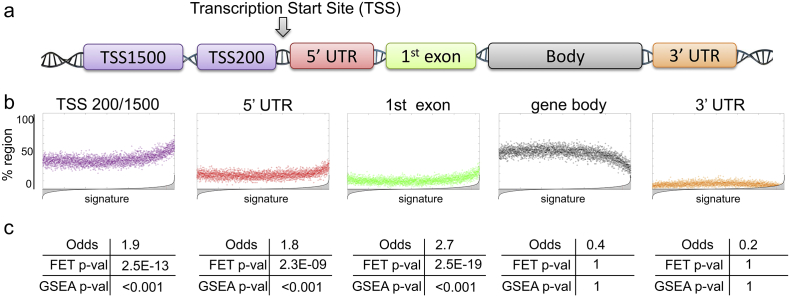
Fig. 3.
Methylation sites from TSS 200/1500, 5′UTR, and 1st exon regions are enriched in methylation signature of ADT resistance. (a) Schematic diagram showing methylation regions profiled on the HumanMethylation450 array. (b) Scatter plots depicting enrichment of each region (i.e., site location) in the differential methylation signature. A reference methylation signature (as in Fig. 2c, ranked from the least differentially methylated (left) to the most differentially methylated (right)) is divided into 100 site-long steps and contribution of each region is calculated as % region inside each step. (c) Odds, Fisher Exact Test (FET) and Gene Set Enrichment Analysis (GSEA) demonstrate statistical significance of the region enrichment from Fig. 3b. For odds and FET, 500 most differentially methylated sites were considered for significance testing. For GSEA, differential methylation signature (Fig. 2c) was utilized as a reference and sites from each region were utilized as query sets. P-value was estimated with 1000 site permutations.