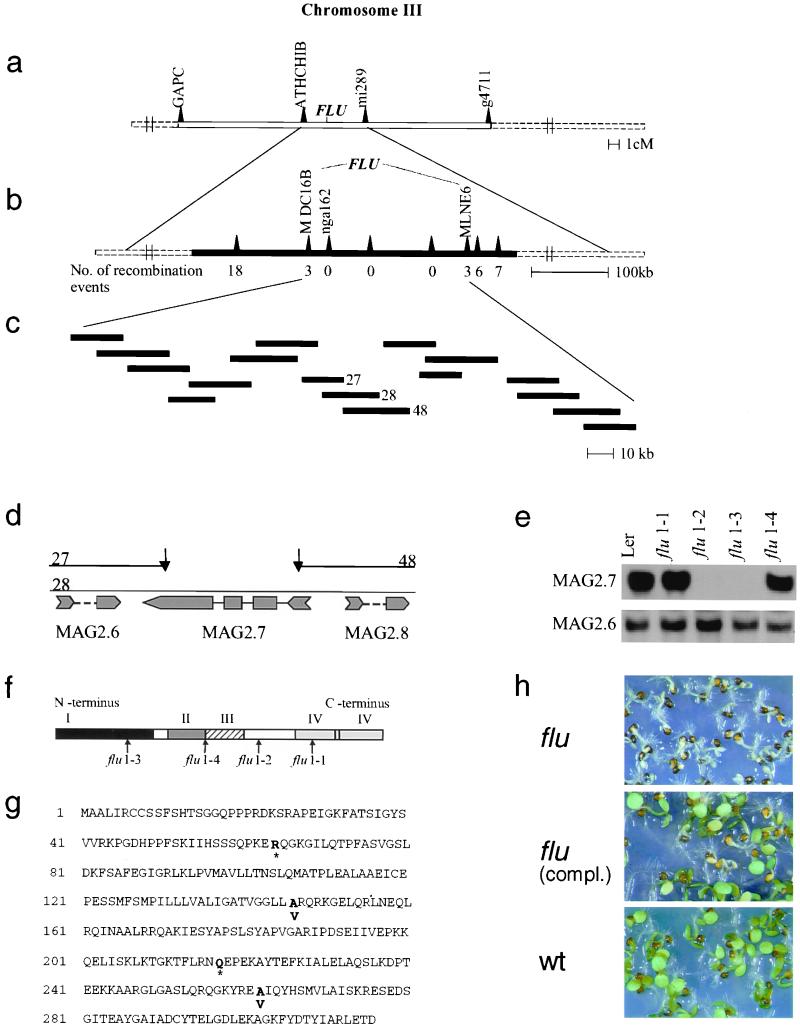
Figure 2.
Identification of the FLU gene. Genetic (a) and physical (b) map of the DNA region on chromosome III of A. thaliana that contains the FLU gene. (c) The region between the markers MDC16B and MLNE6 was encompassed by 17 partially overlapping cosmid clones that were used for the complementation test. Four bacterial artificial chromosome (BAC) clones fully covered this region containing the FLU gene. (d) Details of the physical map of the nonoverlapping part of the cosmid clones 27, 28, and 48. Three ORFs, MAG 2.6, MAG 2.7, and MAG 2.8, have been deduced by the Kazusa DNA Research Institute. (e) Northern blot analysis of RNA extracted from wt (Ler) and the four allelic flu mutants 1–1, -2, -3, -4 grown under continuous white light. Gene-specific probes of MAG 2.6, -2.7, and -2.8 were used for hybridization. Transcripts of MAG 2.8 were not detectable (data not shown). (f) A schematic presentation of the structure of the FLU protein with four domains (I-IV): I, a putative chloroplast signal peptide; II, a hydrophobic region; III, a coiled coil motif; and IV, a TPR region with two TPR motives. Arrows indicate the locations of mutations in the four allelic flu mutants. The extent of the putative chloroplast signal peptide was deduced from the difference in apparent molecular mass of the FLU precursor protein and its mature form that was present inside the plastid (see also Fig. 3). (g) The derived amino acid sequence of FLU deduced from the FLU cDNA sequence. Bold letters indicate positions of mutational changes of the ORF that lead to the premature termination of the polypeptide chain (*) or to amino acid exchanges. Single base pair exchanges in the four allelic flu mutants were found by direct sequencing of PCR products amplified from genomic DNA. (h) The identification of the FLU gene was confirmed by the complementation of the flu mutant with a genomic fragment of wt DNA containing MAG 2.7.