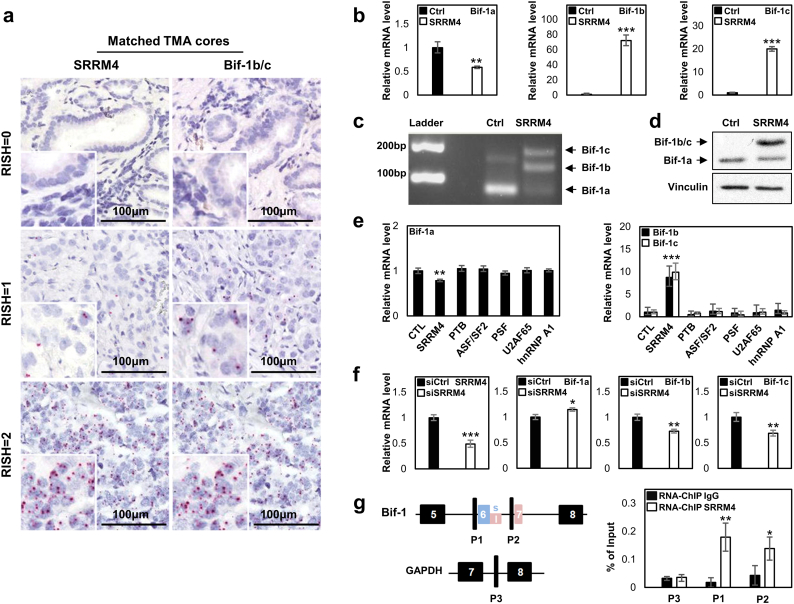
Fig. 4.
SRRM4 regulates alternative RNA splicing of the Bif-1 gene.
(a) Matched tissue cores detected SRRM4 and neural-specific Bif-1 variants in PCa tumor samples.
(b) Real-time qPCR measured mRNA levels of Bif-1 variants in DU145 cells stably expressing control (Ctrl) or SRRM4.
(c) A representative image showed results from regular PCR to detect all Bif-1 variants in DU145 cells stably expressing Ctrl or SRRM4.
(d) Protein lyses from DU145 cells stably expressing Ctrl or SRRM4 were used to measure Bif-1 protein levels by immunoblotting assays.
(e) LNCaP cells were transiently transfected with six RNA splicing factors. Total RNA was extracted to measure the mRNA levels of Bif-1 variants by real-time PCR.
(f) VCaP cells with SRRM4 endogenous expression were transfected with Ctrl or SRRM4 siRNA for 48 h. Total RNA was extracted to measure the mRNA levels of SRRM4 and Bif-1 variants by real-time PCR.
(g) LNCaP cells were transfected with Flag-SRRM4 plasmids. In vivo RNA binding assays were performed using Flag antibody. Eluted RNA fragments were used as templates to perform real-time PCR to measure SRRM4 recruitment to the indicated region. Experiments were repeated three times, and one-way ANOVA or unpaired t-test were performed with *denotes p < 0.05, **denotes p < 0.01 and ***denotes p < 0.001.