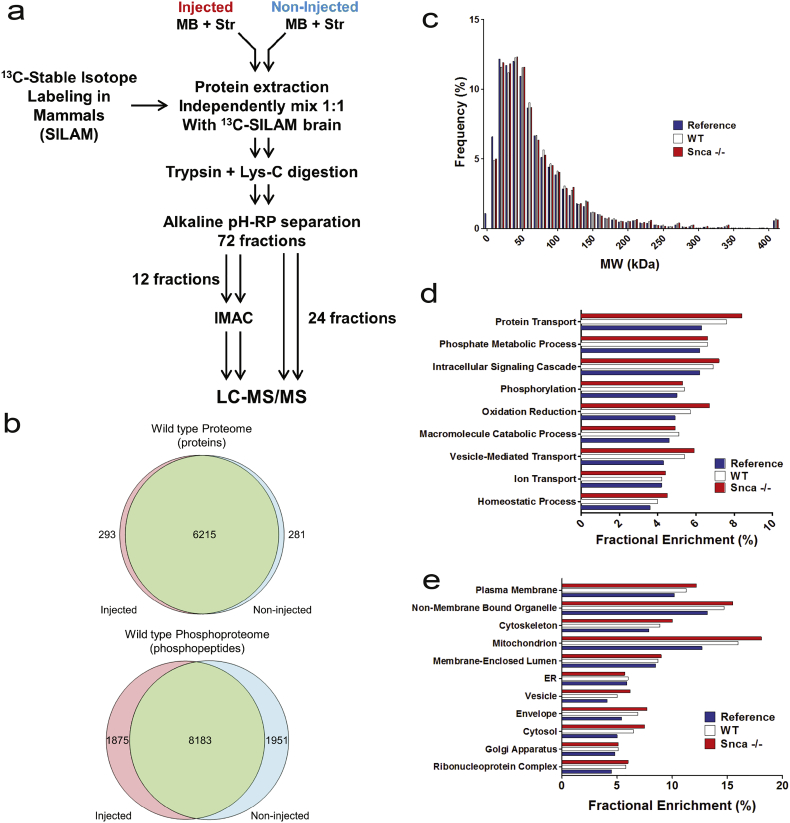
Fig. 1.
Workflow for the acquisition and overall description of proteomes. (a) Overview of proteomic workflow. The midbrain and striatum from the brains of two mice were dissected and combined to generate one sample replicate. The ipsilateral injected and the contralateral non-injected sides were kept separate and processed throughout the workflow independently. Immobilized metal affinity chromatography (IMAC) was used to enrich for phosphorylated peptides. (b) Venn diagrams depicting the number of proteins and phosphopeptides quantified in 4 independent sample replicates in the injected side and non-injected side in wild type mice. The acquired proteomes in wild type and Snca−/− showed typical distribution in terms of molecular weight, cellular location and major biological processes when compared to a whole mouse brain proteome: (b) Distribution of the proteins quantified in wild type and Snca−/− mice based on protein molecular weight. (d) Gene ontology enrichment analysis using cellular components (e) Gene enrichment using biological process and KEGG pathways.