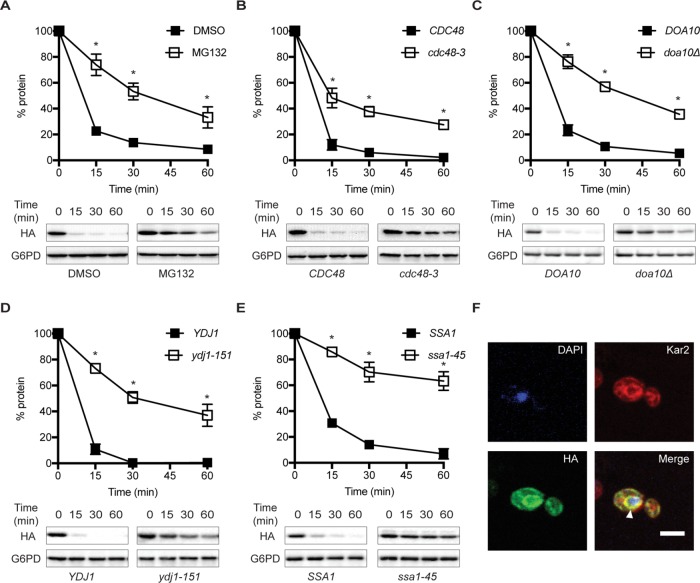
FIGURE 2:
SZ* is only partially degraded by the ERAD pathway. The stabilities of SZ* in the indicated wild-type and mutant strains were determined by cyloheximide chase analyses. (A) The yeast proteasome was inhibited with 100 μM MG132 in a strain lacking PDR5. (B) A strain containing a temperature-sensitive mutation in the AAA-ATPase Cdc48 (cdc48-3) was used. (C) Yeast lacking the ER-associated E3 ligase, Doa10, were examined. (D, E) Cells with a temperature-sensitive mutation in the cytosolic Hsp40, Ydj1 (ydj1-151), and the cytosolic Hsp70, Ssa1, (ssa1-45), were examined. Data represent means ± SE of three to six independent experiments; *p < 0.05. (F) The cellular localization of SZ* was determined by indirect immunofluorescence microscopy. SZ*, ER, and the nucleus were detected with anti-HA antibody, anti-Kar2 antiserum, and DAPI, respectively. Arrowhead denotes the ER. Scale bar: 5 μm.