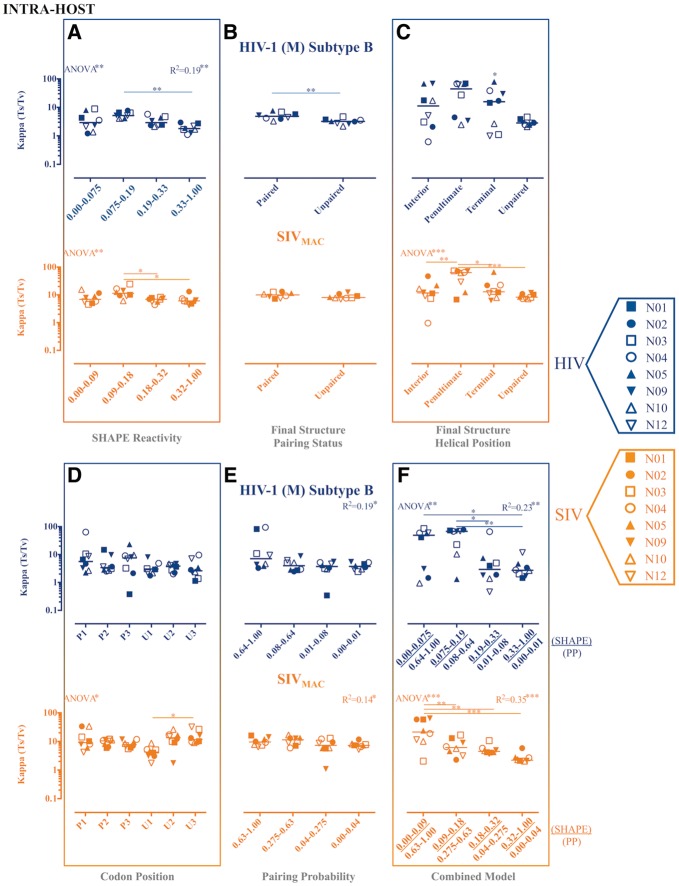
Figure 3.
Maximum likelihood estimates of transition transversion rate ratios (Ts/Tv) for partitioned intra-host HIV-1 (subtype B) and SIVMAC gp120 RNA sequences. Maximum likelihood estimation of the kappa parameter, or Ts/Tv, from each of eight HIV-1-infected patients or SIV-infected macaques was performed in HYPHY (Pond et al. 2005) for internal branches of a fixed phylogeny. Individual subject-specific maximum clade credibility trees used as the fixed tree (topology and branch lengths scaled in time) were obtained from the posterior distribution of a Bayesian analysis of the same data sets. Ts/Tv were estimated for individual site partitions according to SHAPE reactivity (A), final structure pairing status (B), helical position within the final structure (C) codon positions (1, 2, and 3) within paired (P) and unpaired (U) regions according to the final structure (D), pairing probabilities (Pprob) (E), and the nucleotides identified as concordant between SHAPE reactivity and Pprob (F). Error bars represent ±1 SD. *P value < 0.05, **P value < 0.01, ***P value < 0.001 using one-way ANOVA with post-test multiple comparisons (all) and linear trend (A, E, and F). SHAPE reactivity values were obtained from Pollom et al. (2013), whereas Pprob were obtained from Watts et al. (HIV) (2009) and Pollom et al. (SIV) (2013). The final structure referred to in the graph was derived by Pollom et al. (2013) using SHAPE reactivity constraints in RNAstructure (Mathews 2014) thermodynamic folding.