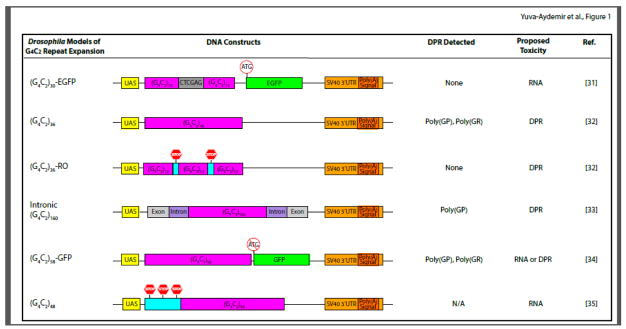
Figure 1. Schematic Representation of Different Drosophila Models of G4C2 Repeat Expansion.
Major features of the DNA constructs used for each model are presented. In all Drosophila G4C2 models, the UAS-GAL4 system is used to overexpress G4C2 repeats with different lengthes, and these models share common SV40 3′UTR containing a polyadenylation signal. In the (G4C2)30-EGFP model, repeats are interrupted in the middle by a six base pair sequence and followed by the EGFP coding region containing the ATG initiation codon. The (G4C2)36 construct contains uninterrupted repeats, while the (G4C2)36-RO construct harbors stop codons between every 12 repeats. The intronic (G4C2)160 construct mimics the human C9ORF72 locus, as 160 copies of repeat sequence are flanked by C9ORF72 intronic sequences and adjacent exons. In the (G4C2)58-GFP model, 58 copies of repeats are upstream to the GFP codon region but without the ATG start codon. The (G4C2)48 construct contains stop codons in each frame 5′ to the repeat sequence. The three columns on the right list DPR proteins detected in these models when phenotypes are observed; the proposed mechanism of toxicity; and the respective references.