Abstract
In this manuscript, a multiple enterocin-producing Enterococcus lactis strain named 4CP3 was used to control the proliferation of Listeria monocytogenes in refrigerated raw beef meat model. Also, the intraspecific genetic differentiation of 4CP3 strain was assessed by Random Amplified Polymorphic DNA Polymerase Chain Reaction (RAPD-PCR) analysis. E. lactis 4CP3 strain was found to produce the enterocins A, B, and P. It displayed activity against L. monocytogenes EGDe 107776 by agar-well diffusion method. The application of E. lactis 4CP3 culture at 107 CFU/g in raw beef meat was evaluated using both ANOVA and ANCOVA linear models in order to examine its effect on the growth of the pathogen L. monocytogenes during refrigerated storage. Hence, a very interesting result in decreasing (P<0.05) and suppressing the growth of L. monocytogenes in refrigerated raw beef meat was shown during 28 days of storage. In conclusion, E. lactis 4CP3 strain might be useful for prevention of the proliferation and survival of L. monocytogenes in raw meat during refrigerated storage.
1. Introduction
Contamination and growth of Listeria monocytogenes in raw beef meats during refrigerated storage have been intensively reported [1–4]. Effectively, L. monocytogenes is known to be a major concern for the meat processing industry causing listeriosis in humans [4, 5]. This fact constitutes a significant public health issue [6]. Indeed, this virulent foodborne pathogen is psychrophile which is able to grow at temperatures as low as 0°C, adapted to several food systems and the contaminated foods do not present unusual odour, texture, or taste which evade control in human foodstuffs and increase its danger in products [7]. In this context, many researches were performed in order to develop natural agents other than antibiotics and chemically synthesised additives to ensure the safety and maintain the security of foods as public health issues [8]. Among these natural agents, lactic acid bacteria (LAB) have received great attention in terms of food safety and are mainly used in foods for different technological effects because of their potent Generally Recognised as Safe (GRAS) status [9]. In fact, LAB are implicated in the biopreservation and prolongation of the shelf-life of diverse food products owing to their production of antimicrobial substances [10, 11].
Bacteriocins are among the most studied antimicrobial substances produced by LAB [12]. These antimicrobial peptides (bacteriocins) may be added as biopreservatives to improve the microbial stability and safety of chill-stored fresh meat [13, 14]. Among the studied bacteriocins in meat and meat products we can cite the nisin. Produced by Lactococcus lactis, nisin was used successfully as food preservative in more than 50 countries [4]. This purified bacteriocin, nisin, showed bactericidal effect against Listeria monocytogenes in fresh meat and its application at 500 IU/ml engenders a significant reduction in the L. monocytogenes in meat [4]. On the other hand, direct use of bacteriocin-producing cells is one of the most practical strategies that seem to be more feasible from an economic point of view and lesser legal restrictions compared to the direct addition of purified bacteriocins. This can benefit the food industry in terms of microbiological quality and safety as well as cost since it reduces food losses caused by microbiological spoilage. Enterococci, isolated from diverse food sources, are among the most evaluated LAB as protective cultures in different foods due to their produced bacteriocins that are able to inhibit several key foodborne pathogens such as L. monocytogenes [15, 16]. Effectively, there are many strains of Enterococcus spp. that have been applied to the control of L. monocytogenes in different food systems [17–19]. Nowadays, advanced technologies have been developed for starters and protective cultures to enhance their efficacy and applicability in food products such as bioactive packaging and encapsulation [20]. Even though enterococci have been found naturally in different types of foods, their use in food products is controversial because they are considered as opportunistic pathogens implicated in several nosocomial infections and constitute a source of multiple antibiotic resistances [16].
The objectives of this work were to characterise genetically the multiple enterocin-producing Enterococcus lactis 4CP3 strain using RAPD-PCR analysis and evaluate its effect on the growth of L. monocytogenes in refrigerated raw beef meat.
2. Material and Methods
2.1. Strains and Growth Conditions
E. lactis 4CP3 strain was isolated from a raw shrimp (Palaemon serratus). The kinetic of bacteriocin production by 4CP3 strain was evaluated in MRS (de Man, Rogosa and Sharpe, Biokar Diagnostics, Beauvais, France) broth under aerobic conditions at 30°C [21]. This isolate was a multiple enterocin-producing strain able to produce the enterocins A, B, and P [21]. Also, it has been shown to display bactericidal mode of action against the pathogenic Gram-positive strain of L. monocytogenes EGDe 107776. It was grown overnight in MRS broth at 30°C.
E. faecium VC185 strain was isolated from Italian cheese [22]. This isolate is a non-bacteriocin-producing strain and is used in this study as the control strain in the meat challenge experimentation. It was also grown in MRS broth.
L. monocytogenes EGDe 107776 strain was used as the indicator strain for antimicrobial activity tests and the target microorganism in the microbiological challenge test. It was grown in BHI (Brain Heart Infusion, Biokar Diagnostics, Beauvais, France) broth and cultured on ALOA (Agar Listeria Ottaviani and Agosti, BIO-RAD, Marnes-la-Coquette, France) medium for enumeration [23].
E. faecium MMT21 strain was isolated from Tunisian rigouta cheese [24]. This isolate is used as the target strain in the direct detection of antimicrobial activity by overlaying with MRS soft agar in order to examine the capacity of E. lactis 4CP3 to produce bacteriocins in beef samples during the challenge test.
2.2. Random Amplified Polymorphic DNA-PCR (RAPD-PCR) Analysis
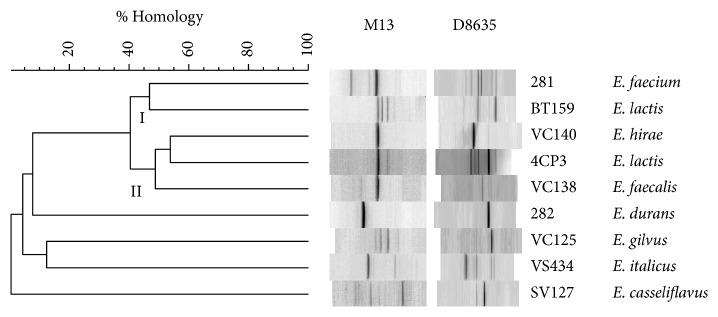
Genomic DNA used for RAPD-PCR amplification was extracted from overnight culture of E. lactis 4CP3 in M17 broth at 30°C according to Cremonesi et al., 2006 [25]. RAPD-PCR amplification was realised using the universal primers M13 and D8635 as described by Andrighetto et al., 2001 [26]. Amplification products were separated by electrophoresis on agarose gel (1.5%) in 1 × TAE buffer at 100 mV for 99 min. The gels were stained in ethidium bromide and photographed on a UV transilluminator. Photo-positives were scanned into a computer and were analysed using the BioNumeric 5.0 software package (Applied Maths NV, Sint-Martens-Latem, Belgium). Grouping of the RAPD-PCR patterns was performed using the Unweighted Pair Group Method with Arithmetic Averages (UPGMA) cluster analysis. The reproducibility value of the RAPD-PCR assay, calculated from two repetitions of independent amplification of type strains, was higher than 90%. The RAPD-PCR profiles obtained with both primers (M13 and D8635) were analysed together to obtain a single dendrogram.
2.3. Antimicrobial Activity against L. monocytogenes
Overnight culture of E. lactis 4CP3 strain incubated at 30°C in MRS broth was centrifuged at 10,000 × g for 10 min to obtain a cell free supernatant which was neutralised at pH 6 with NaOH (1 M) in order to eliminate the inhibitory effect of organic acids, and sterilised by filtration (0.22 µm, Millipore, Bedford, MA) [21]. The antimicrobial activity of the cell free supernatant of E. lactis 4CP3 against L. monocytogenes EGDe 107776 was assayed by the agar-well diffusion method according to Ben Braïek et al., 2017 [27]. The BHI agar plate was incubated at 37°C for 24 h and the diameter of the inhibition zone was measured in millimetres (mm).
2.4. Influence of E. lactis 4CP3 Strain addition on the Growth of L. monocytogenes EGDe 107776 in Raw Beef Meat
2.4.1. Preparation and Inoculation of Raw Beef Samples
Raw beef meat was bought from a local supermarket in the region of Sousse (Tunisia) and transported to the laboratory under refrigerated conditions to be processed immediately. The prepared beef meat was aseptically cut into five equal portions of 200 g each (BF1-BF5). In order to reduce to the lowest possible levels the number of intrinsic microorganisms attached to the surface of beef meat portions, each piece was immersed in boiling sterile water for 5 min [28]. The cooked surface of the meat samples was eliminated with sterile knives under aseptic conditions [28]. These meat portions were further cut into small pieces of about 2 × 2 × 0.5 cm. Prior to beef meat contamination with L. monocytogenes and inoculation with LAB strains, beef portions were examined for any contamination by mesophilic and psychrotrophic flora. Total mesophilic bacteria were determined on plate count agar (PCA; Difco Laboratories, Detroit, MI, USA), incubated at 30°C for 48 h. Psychrotrophic counts were determined as described above for mesophilic bacteria except that the incubation was at 4°C for 7 days [29].
E. lactis 4CP3 strain was grown in MRS broth at 30°C for 24 h to reach the maximum of its bacteriocin production (1400 AU/ml) [21]. L. monocytogenes EGDe 107776 was subcultured in BHI broth firstly at 37°C for 18 h to reach the early stationary phase with cells at the same physiological state, then at 10°C (temperature of the meat storage) for 3 days as adaptation step to the storage conditions. The in situ influence of the application of E. lactis 4CP3 strain on the survival of L. monocytogenes EGDe 107776 in raw beef meat was assessed according to the slightly modified method of Dortu et al., 2008 [6]. Briefly, the portions BF2 and BF3 were firstly surface inoculated at 107 CFU/g of meat with E. lactis 4CP3 and E. faecium VC185 strains, respectively. After absorption of the LAB inocula at room temperature, the BF1, BF2, and BF3 meat portions were surface contaminated with 105 CFU of L. monocytogenes EGDe 107776/g of meat. A sterile spreader was used to distribute homogeneously the inocula. The portion BF1 served as control (artificially contaminated only with L. monocytogenes EGDe 107776). The portion BF4 and BF5 were not contaminated with L. monocytogenes EGDe 107776 but were inoculated only with 4CP3 and VC185 strains, respectively, at 107 CFU/g of meat.
2.4.2. Storage and Enumerations
The raw beef meat portions were placed separately in sterile plastic bags and stored for 28 days at 10°C. The choice of this storage temperature relies firstly on the growth conditions of enterococcal strains used in this study that are not able to grow at temperatures lower than 10°C [30]. Secondly, meat storage at 10°C aimed to mimic the worst-case scenario for cold storage according to Kennedy et al., 2005 [31].
L. monocytogenes EGDe 107776 plate counts were determined on ALOA agar plates according to NF EN ISO 11290-2: 2005 [23]. The LAB counts were determined on MRS agar plates after incubation at 30°C for 24 h. Microbial enumerations were expressed as log10 CFU/g of beef meat. Plates containing 25-250 colonies were selected and counted, and the average number of CFU/g was calculated. These cell counts were performed every 6 h during 48 h, every day up to day 7 and every 7 days until day 28.
To detect enterocin production by E. lactis 4CP3 in raw beef meat, homogenates from the portions BF2 and BF4 stored at days 0, 7, 14, 21, and 28 were plated on MRS agar. After aerobic incubation at 30°C for 24 h, the plates were further overlaid with the indicator strain E. faecium MMT21 in soft agar and incubated overnight at 30°C. Bacteriocin production was indicated by clear inhibition zones around the colonies.
2.5. Statistical Analysis
Measurements were carried out in triplicates and repeated three times. A one-way analysis of variance (ANOVA) was applied for each parameter by using SPSS 19 statistical package (SPSS Ltd., Woking, UK). Means and standard errors were calculated and a probability level of P<0.05 was used in testing the statistical significance of all experimental data. Tukey's post hoc test was used to determine significance of mean values for multiple comparison at P<0.05. On the other hand, we used linear mixed models assuming the error to compare the CFU values among treatments with different days. Mixed models were fitted using SPSS 19 and followed by post hoc contrasts through the origin. The interpretation of the statistical output of a mixed model requires an understanding of how to explain the relationships among the fixed and random effects in terms of the hierarchy levels.
3. Results and Discussion
3.1. RAPD-PCR Analysis
E. lactis 4CP3 strain was previously identified by different genetic methods: 16S rRNA gene sequencing, rpoA and pheS gene sequencing, and 16S-23S rRNA intergenic spacer analysis (RSA) [21]. Indeed, RSA analysis demonstrated that E. lactis 4CP3 strain displayed the same 16S-23S profile as the type strain E. lactis DSM 23655T (BT159), while in this study, they presented different RAPD-PCR patterns as shown in Figure 1. Accordingly, two clusters (I and II) could be detected at a similarity level of 45% arbitrarily chosen for defining species. Interestingly, E. lactis 4CP3 and the type strain E. lactis BT159 were found to belong to different clusters even though they belong to the same species (Figure 1). This genetic differentiation between E. lactis 4CP3 and BT159 strains as illustrated by their clustering in the dendrogram could be related to their different isolation sources. Effectively, our E. lactis 4CP3 strain was isolated from a fresh shrimp sample of Palaemon serratus [21], while, E. lactis BT159 strain was isolated from an Italian cheese sample [32]. Therefore, RAPD-PCR analysis constitutes a rapid molecular method that could detect genetic diversity at a strain level with accuracy.
Figure 1.
Unweighted pair group method using arithmetic averages (UPGMA) based dendrogram derived from the combined RAPD-PCR profiles generated with primers M13 and D8635 of E. lactis 4CP3 strain, type strains, and other enterococcal strains. The type strains used in this analysis were E. lactis DSM 23655T (BT159), E. faecium DSM 20477T (281), and E. durans DSM 20633T (282) from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig (Germany). The other enterococcal strains were E. gilvus (VC125), E. italicus (VS434), E. hirae (VC140), E. faecalis (VC138), and E. casseliflavus (SV127) from the bacterial collection of ISPA-CNR (Milan, Italy).
3.2. Antilisterial Activity
In vitro antibacterial assay of E. lactis 4CP3 strain showed high antilisterial activity (P<0.05) against L. monocytogenes EGDe 107776 with a clear growth inhibition zone diameter of 12 mm on BHI agar (Table 1). This result corroborates previous finding described for E. faecium strains [17]. This antagonistic activity towards L. monocytogenes was due to the production of the enterocins (A, B, and P) as previously demonstrated by Ben Braïek et al., 2018 [21]. In fact, the enterocins A, B, and P are among the most characterised bacteriocins and are known to be active against Listeria spp. as reported by Vandera et al. 2017 [16] and Rehaiem et al. 2014 [33].
Table 1.
Inhibitory spectrum of E. lactis 4CP3, CR4, CL, 5CP2, C15, and C23 strains against L. monocytogenes EGDe 107776.
| Test strain | Diameter of inhibition zones (mm) |
|---|---|
| E. lactis 4CP3 | 12.00±1.00d |
| E. lactis CR4 | 10.00±1.00c |
| E. lactis CL | 5.00±0.00b |
| E. lactis 5CP2 | 5.00±0.00b |
| E. lactis C15 | 0.00±0.00a |
| E. lactis C23 | 0.00±0.00a |
| PC | 18.00±2.00e |
| NC | 0.00±0.00a |
Results are reported as means ± standard error of three replicates.a–e: averages with different letters in the same column, for each diameter of inhibition zones, are significantly different (P<0.05). PC: positive control (Novobiocin 1 mg/ml) and NC: negative control (noninoculated MRS broth medium).
3.3. Influence of E. lactis 4CP3 Strain addition on the Growth of L. monocytogenes EGDe 107776 in Raw Beef Meat Using ANOVA
Meat is considered to be one of the most frequently contaminated foods with L. monocytogenes [3]. According to Rapid Alert System for Food and Feed in 2016 [34], 20% of L. monocytogenes notifications were due to the contamination of meats other than poultry. In this context, a challenge test to control the growth of L. monocytogenes in raw beef meat inoculated with an enterocin-producing E. lactis strain was carried out. Furthermore, it is important to mention that high levels of intrinsic nonpathogenic microorganisms may have an inhibitory effect on pathogens present in meat by outcompeting them [28]. For this reason, our meat samples were subjected to boiling treatment with sterile water as described above in order to reduce the number of factors that could be implicated in the listerial growth in beef food models and to ovoid interferences of colonies on plating agar.
It should be noted that the analysis of mesophilic and psychrotrophic bacteria from meats treated separately with E. lactis 4CP3 and E. faecium VC185 strains showed an inhibition of these bacteria (mesophilic and psychrotrophic) since only the LAB, E. lactis and E. faecium, were identified (data not shown). In fact, the microbial load of aerobic mesophilic plate count and psychrotrophic count was zero, demonstrating the effective process of the boiling sterile water immersion intervention to eliminate these bacteria (aerobic mesophilic and psychrotrophic) from meat portions.
As demonstrated by Figure 2, there were no significant differences (P>0.05) in the growth of E. lactis 4CP3 strain and E. faecium VC185 strain in raw beef meat showing comparable growth rates increasing by 3.43 logs and 3.35 logs, respectively, in 28 days of storage. The population of L. monocytogenes EGDe 107776 in portion BF1 (positive control: artificially contaminated with 105 CFU/g of meat) underwent an increase from 105 CFU/g to 2.87×109 CFU/g after 28 days (Figure 3).
Figure 2.
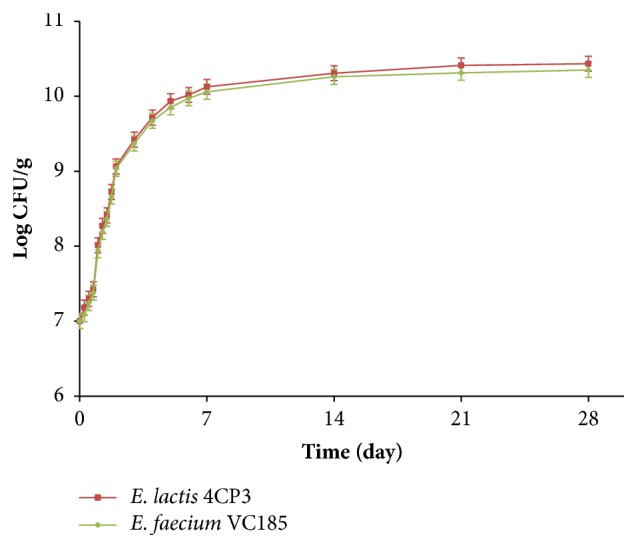
Growth of LAB strains in raw beef meat. Red square: E. lactis 4CP3 (enterocin-producing LAB strain) and green diamond: E. faecium VC185 (non-bacteriocin-producing LAB strain).
Figure 3.
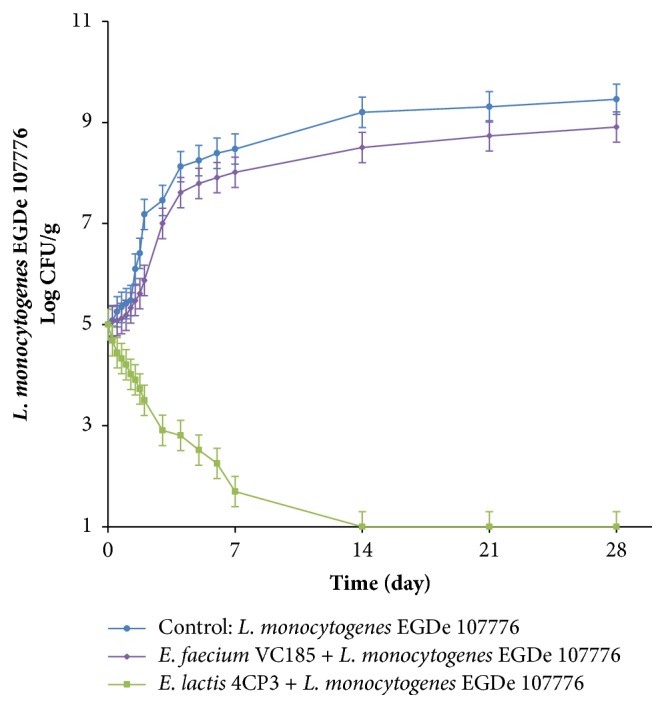
Influence of inhibitory LAB cultures on the growth of L. monocytogenes EGDe 107776 in raw beef meat during storage at 10°C. Blue circle: control (L. monocytogenes EGDe 107776 without enterocin-producing LAB strain), violet diamond: E. faecium VC185 (non-bacteriocin-producing LAB strain), and green square: E. lactis 4CP3 strain (enterocins A, B and P-producing strain).
Statistical evaluation of the data relating to the growth behaviour of L. monocytogenes EGDe 107776 in raw beef meat inoculated with E. lactis 4CP3 strain showed significant reduction (P<0.05) of listerial population by 6.77 log units compared with the untreated control after 7 days of storage (Figure 3). Then, the growth of L. monocytogenes was completely inhibited from day 14 to the end of the experiment.
The application of the non-bacteriocin-producing E. faecium VC185 strain led to a very low reduction of L. monocytogenes populations. These counts were only 0.46 log units and 0.55 log units lower than the control counts after 7 and 28 days of storage, respectively. Moreover, no significant growth (P>0.05) of L. monocytogenes EGDe 107776 was observed in the portions BF4 and BF5 which were only inoculated with LAB strains at 107 CFU/g and not contaminated with the listerial pathogen.
3.4. In Situ Detection of Enterocin Production in Raw Beef Meat
Overlay assays with MRS agar plates were realised in order to detect in situ production of enterocins by E. lactis 4CP3 strain in beef meat samples during the refrigerated storage period. After incubation, enterocin production was indicated by observation of obvious inhibition zones around the colonies grown on MRS agar medium. Generally, it was shown that the application of the multiple enterocin-producing E. lactis 4CP3 strain in raw beef meat led to a greater (P<0.05) inhibition of L. monocytogenes EGDe 107776 than that of the non-bacteriocin-producing E. faecium VC185 strain (Figure 3). Also, it was demonstrated in this study that this enterococcal culture strongly (P<0.05) inhibited the growth of L. monocytogenes in beef meat after the first 7 days of the challenge test and then suppressed dramatically the pathogen. This potent inhibitory behaviour of E. lactis 4CP3 towards L. monocytogenes could be explained by the enterocin production as confirmed above. In fact, enterocins A and P have strong antilisterial activity against L. monocytogenes; however, enterocin B displays synergistic activity with enterocin A [16, 33]. Thus, our present results corroborate these previous findings indicating that enterocins A and B may synergistically inhibit L. monocytogenes growth. Likewise, a synergistic interaction between the three produced enterocins (A, B, and P) by E. lactis 4CP3 could be proposed reflecting thus its effectiveness in raw beef meat preservation. Similar results reporting the biocontrol of L. monocytogenes in different meat products with bacteriocinogenic LAB were previously described by Dortu et al., 2008 [6], Pragalaki et al., 2013 [35], and Giello et al., 2018 [36]. Therefore, it is clear that application of bacteriocin-producing LAB in meats and meat products have been attracting considerable interest as alternative natural food preservatives to extend shelf-life and safety of meats these recent years [37, 38]. Effectively, direct application of bacteriocin-producing LAB is among the most advanced and practical approaches from economic and regulatory status point of views. Indeed, this bacterial use does not need many processing steps such as purification and has fewer legal restrictions and limits compared to the direct application of purified bacteriocins [39].
3.5. Influence of E. lactis 4CP3 Strain addition on the Growth of L. monocytogenes EGDe 107776 in Raw Beef Meat Using General Linear Model (ANCOVA)
Analysis of covariance (ANCOVA) is a general linear model which blends ANOVA and regression. ANCOVA evaluates whether the means of dependent variables (11 sampling days: 0, 1, 2, 3, 4, 5, 6, 7, 14, 21, and 28 days of storage at 10°C) which are equal across levels of categorical independent variables (five trials: Trial 1: BF1, Trial 2: BF2, Trial 3: BF3, Trial 4: BF4, and Trial 5: BF5) and inversely. In order to simplify the obtained results, for each meat product, firstly (i) we analysed parameters between 0 and 7 days and secondly (ii) all parameters were evaluated between 7 and 28 days.
3.5.1. ANCOVA Parameter Analyses between 0 and 7 Days
As in ANCOVA, writing out the full regression model and then simplifying tells us that the intercept for day zero was 4.000 (4.194436–0.194436) and this was lower than log10 CFU at the seventh day group (t= -0.053). Similarly, we knew that the days 0, 1, 2, 3, 4, 5, and 6 had lower intercepts than the 7th day. The trial coefficient of 1.160110 represented the average for each subsequent trial for the baseline on day 7. The interaction estimates tell the difference in slope for other day groups compared to the seventh day groups (Table 2(a)). We are particularly interested in the conclusion that we are 95% confident that the control sample had an effect on the CFU that was between 16.611213 points more and -17.731432 points less than treatment for beef meat (Table 2(a)).
Table 2.
(a) Raw beef meat estimates of trials fixed effects between 0 and 7 days.
| Parameter | Estimate | Std. Error | Df | t | Sig. | 95% Confidence Interval | |
|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||
| Intercept | 4.194436 | 2.595846 | 24 | 1.616 | 0.119 (ns) | -1.163127 | 9.551998 |
| Day 0 | -0.194436 | 3.671080 | 24 | -0.053 | 0.958 (ns) | -7.771173 | 7.382302 |
| Day 1 | -0.699838 | 3.671080 | 24 | -0.191 | 0.850 (ns) | -8.276576 | 6.876899 |
| Day 2 | -0.041189 | 3.671080 | 24 | -0.011 | 0.991 (ns) | -7.617927 | 7.535548 |
| Day 3 | -0.064947 | 3.671080 | 24 | -0.018 | 0.986 (ns) | -7.641685 | 7.511790 |
| Day 4 | 0.392455 | 3.671080 | 24 | 0.107 | 0.916 (ns) | -7.184282 | 7.969193 |
| Day 5 | 0.285756 | 3.671080 | 24 | 0.078 | 0.939 (ns) | -7.290981 | 7.862494 |
| Day 6 | 0.235276 | 3.671080 | 24 | 0.064 | 0.949 (ns) | -7.341462 | 7.812013 |
| Day 7 | 0a | 0 | . | . | . | . | . |
| Trial | 1.160110 | 6.194913 | 94193.706 | 0.187 | 0.851 (ns) | -10.981849 | 13.302068 |
| Day 0 × Trial | -0.560110 | 8.760930 | 94193.706 | -0.064 | 0.949 (ns) | -17.731432 | 16.611213 |
| Day 1 × Trial | -0.273876 | 8.760930 | 94193.706 | -0.031 | 0.975 (ns) | -17.445198 | 16.897447 |
| Day 2 × Trial | -0.233863 | 8.760930 | 94193.706 | -0.027 | 0.979 (ns) | -17.405185 | 16.937460 |
| Day 3 × Trial | -0.126228 | 8.760930 | 94193.706 | -0.014 | 0.989 (ns) | -17.297550 | 17.045095 |
| Day 4 × Trial | -0.160129 | 8.760930 | 94193.706 | -0.018 | 0.985 (ns) | -17.331451 | 17.011194 |
| Day 5 × Trial | -0.097362 | 8.760930 | 94193.706 | -0.011 | 0.991 (ns) | -17.268685 | 17.073960 |
| Day 6 × Trial | -0.067141 | 8.760930 | 94193.706 | -0.008 | 0.994 (ns) | -17.238464 | 17.104181 |
| Day 7 × Trial | 0a | 0 | . | . | . | . | . |
a : this parameter is set to zero because it is redundant. Std. Error: standard error, df: the degrees of freedom, t: Student's t-statistic, and Sig.: the p-value (associated with the correlation). ns: P>0.05.
(b) Raw beef meat estimates of days fixed effects.
| Parameter | Estimate | Std. Error | df | t | Sig. | 95% Confidence Interval | |
|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||
| Intercept | 7.467589 | 0.246726 | 26.643 | 30.267 | 0.000 (∗∗∗) | 6.961032 | 7.974147 |
| Trial 1 | -2.190545 | 0.334916 | 23.708 | -6.541 | 0.000 (∗∗∗) | -2.882229 | -1.498862 |
| Trial 2 | -3.072563 | 0.334916 | 23.708 | -9.174 | 0.000 (∗∗∗) | -3.764247 | -2.380879 |
| Trial 3 | -2.595970 | 0.334916 | 23.708 | -7.751 | 0.000 (∗∗∗) | -3.287653 | -1.904286 |
| Trial 4 | 0.028207 | 0.334916 | 23.708 | 0.084 | 0.934 (ns) | -0.663477 | 0.719891 |
| Trial 5 | 0a | 0 | . | . | . | . | . |
| Day | 0.489768 | 0.070900 | 21.369 | 6.908 | 0.000 (∗∗∗) | 0.342479 | 0.637057 |
| Trial 1 × Day | 0.103902 | 0.080060 | 23.708 | 1.298 | 0.207 (ns) | -0.061443 | 0.269246 |
| Trial 2 × Day | -0.836490 | 0.080060 | 23.708 | -10.448 | 0.000 (∗∗∗) | -1.001834 | -0.671146 |
| Trial 3 × Day | 0.080347 | 0.080060 | 23.708 | 1.004 | 0.326 (ns) | -0.084998 | 0.245691 |
| Trial 4 × Day | 0.005842 | 0.080060 | 23.708 | 0.073 | 0.942 (ns) | -0.159502 | 0.171186 |
| Trial 5 × Day | 0a | 0 | . | . | . | . | . |
a : this parameter is set to zero because it is redundant. Std. Error: standard error, df: the degrees of freedom, t: Student's t-statistic, and Sig.: the p-value (associated with the correlation). Trial 1: BF1 (control sample), Trial 2: BF2, Trial 3: BF3, Trial 4: BF4, and Trial 5: BF5. ns: P>0.05, ∗∗∗: P<0.001.
Equally, ANCOVA indicated that there were no statistically significant differences (P>0.05) among the treatments between 0 and 7 days (Table 2(a)).
As shown in Table 2(b), writing out the full regression model then simplifying tells us that the intercept for trial 1 was 5.277044 (7.467589–2.190545). Similarly, we knew the trials 2, 3, 4, and 5. The day coefficient of 0.489768 represented the average for each subsequent trial for the baseline on the trial 5 (Table 2(b)). The interaction estimates tell the difference in slope for other trial groups compared to the fifth groups (Table 2(b)).
The treatments BF1 (control sample), BF3 (E. faecium VC185 strain at 107 CFU/g of meat + 105 CFU of L. monocytogenes EGDe 107776/g of meat), BF4 (only E. lactis 4CP3 strain at 107 CFU/g of meat), and BF5 (only E. faecium VC185 strain at 107 CFU/g of meat) had no significant differences (P>0.05) between them. However, at the P<0.001 confidence level, the treatment of E. lactis 4CP3 strain at 107 CFU/g of meat + 105 CFU of L. monocytogenes EGDe 107776/g of meat (BF2) was statistically different and was more sensitive to dose than the other trials (Table 2(b)).
It is very important to realise that the parameter estimates given in the fixed effects were estimates of mean parameters. The covariance parameters are presented in Table 3. Equally, the intercepts' variances were estimated as 0.134603 and 6.125832 (Table 3). The null hypothesis for this parameter was a variance of zero, which would indicate that a random effect was not needed. The statistical test is called Wald Z statistic. On the other hand, the hypothesis (Wald Z = 0.000, P = 1.00) was accepted and the null hypothesis (Wald Z = 1.067, P = 0.286) was rejected. In fact, we conclude that we do need a random intercept (Table 3). This suggests that there are important unmeasured explanatory variables for each subject that raise or lower their performance in a way that appears random because we do not know the values of the missing explanatory variables.
Table 3.
Estimates of covariance parameters in raw beef meat samples between 0 and 7 days.
| Parameter | Estimate | Wald Z | Sig. | |
|---|---|---|---|---|
| Residual | 0.134603 | 3.443 | 0.001 | |
| Day [subject = id] | Variance | 0.006163 | 1.067 | 0.286 |
| Residual | 6.125832 | 3.464 | 0.001 | |
| Trial [subject = id] | Variance | 37.764364a | . | . |
a : this covariance parameter is redundant. The test statistic and confidence interval cannot be computed. Sig.: the p-value (associated with the correlation).
3.5.2. ANCOVA Parameter Analyses between 7 and 28 Days
For a period ranged between 7 and 28 days of storage, the ANCOVA intercept for day seven was 4.194435 (4.664446–0.470011) and this was lower than log10 CFU at the twenty-eighth day group (t= -0.081) (Table 4(a)). Similarly, the days 7, 14, and 21 had lower intercepts than the day 28. The trial coefficient of 1.121930 represented the average for each subsequent trial for the baseline on the day 28 (Table 4(a)). Furthermore, the treatment control sample (Trial 1) had an effect on the CFU (Table 4(a)).
Table 4.
(a) Raw beef meat estimates of trials fixed effects between 7 and 28 days.
| Parameter | Estimate | Std. Error | Df | t | Sig. | 95% Confidence Interval | |
|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||
| Intercept | 4.664446 | 4.093800 | 12 | 1.139 | 0.277 (ns) | -4.255177 | 13.584069 |
| Day 7 | -0.470011 | 5.789507 | 12 | -0.081 | 0.937 (ns) | -13.084262 | 12.144241 |
| Day 14 | -0.239269 | 5.789507 | 12 | -0.041 | 0.968 (ns) | -12.853521 | 12.374983 |
| Day 21 | -0.138680 | 5.789507 | 12 | -0.024 | 0.981 (ns) | -12.752932 | 12.475572 |
| Day 28 | 0a | 0 | . | . | . | . | . |
| Trial | 1.121930 | 2.383010 | 0.000 | 0.471 | 1.000 (ns) | -16.706240 | 18.950100 |
| Day 7 × Trial | 0.038180 | 3.370085 | 0.000 | 0.011 | 1.000 (ns) | -25.174660 | 25.251019 |
| Day 14 × Trial | 0.021102 | 3.370085 | 0.000 | 0.006 | 1.000 (ns) | -25.191737 | 25.233942 |
| Day 21 × Trial | 0.020506 | 3.370085 | 0.000 | 0.006 | 1.000 (ns) | -25.192333 | 25.233345 |
| Day 28 × Trial | 0a | 0 | . | . | . | . | . |
a : this parameter is set to zero because it is redundant. Std. Error: standard error, df: the degrees of freedom, t: Student's t-statistic, and Sig.: the p-value (associated with the correlation).
ns: P>0.05.
(b) Raw beef meat estimates of days fixed effects.
| Parameter | Estimate | Std. Error | df | t | Sig. | 95% Confidence Interval | |
|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||
| Intercept | 10.015610 | 0.209103 | 10 | 47.898 | 0.000 (∗∗∗) | 9.549700 | 10.481519 |
| Trial 1 | -1.671006 | 0.295716 | 10 | -5.651 | 0.000 (∗∗∗) | -2.329902 | -1.012110 |
| Trial 2 | -8.316640 | 0.295716 | 10 | -28.124 | 0.000 (∗∗∗) | -8.975536 | -7.657744 |
| Trial 3 | -2.204440 | 0.295716 | 10 | -7.455 | 0.000 (∗∗∗) | -2.863336 | -1.545544 |
| Trial 4 | 0.047959 | 0.295716 | 10 | 0.162 | 0.874 (ns) | -0.610937 | 0.706855 |
| Trial 5 | 0a | 0 | . | . | . | . | . |
| Day | 0.013178 | 0.010908 | 10 | 1.208 | 0.255 (ns) | -0.011126 | 0.037482 |
| Trial 1 × Day | 0.030534 | 0.015426 | 10 | 1.979 | 0.046 (∗) | -0.003837 | 0.064905 |
| Trial 2 × Day | -0.043134 | 0.015426 | 10 | -2.796 | 0.009 (∗∗) | -0.077504 | -0.008763 |
| Trial 3 × Day | 0.028453 | 0.015426 | 10 | 1.845 | 0.095 (ns) | -0.005917 | 0.062824 |
| Trial 4 × Day | 0.001487 | 0.015426 | 10 | 0.096 | 0.925 (ns) | -0.032884 | 0.035857 |
| Trial 5 × Day | 0a | 0 | . | . | . | . | . |
a : this parameter is set to zero because it is redundant. Std. Error: standard error, df: the degrees of freedom, t: Student's t-statistic, and Sig.: the p-value (associated with the correlation). Trial 1: BF1 (control sample), Trial 2: BF2, Trial 3: BF3, Trial 4: BF4, and Trial 5: BF5. ns: P>0.05, ∗: P<0.05, ∗∗: P<0.01, and ∗∗∗: P<0.001.
As shown in Table 4(a), there were no significant differences (P>0.05) among the trials and days 7, 14, 21, and 28.
Indeed, the lower and upper bound of the confidence interval for the mean difference ranged from -25.174660 points to 25.251019 points (Table 4(a)). The full regression model then simplifying the intercept for the control sample was 8.344604 (10.015610–1.671006) (Table 4(b)). Similar results were shown for 2, 3, 4, and 5 trial groups. The day coefficient was 0.013178.
The effects of treatments, time, and their interaction on the inhibition of L. monocytogenes are shown in Table 4(b). no significant interaction (P>0.05) between treatments BF3 and BF4, and the time of storage in meat. However, interestingly, at the P<0.01 confidence level, BF2 and time of storage were found to have a highly significant effect regarding inhibition of L. monocytogenes EGDe 107776 in meat (Table 4(b)).
Moreover, the intercepts' variances were estimated as 0.029149 and 15.235632. Besides, the hypothesis (Wald Z = 0.000, P = 1.00) was accepted for beef meat samples (Table 5).
Table 5.
Estimates of covariance parameters in raw beef meat samples between 7 and 28 days.
| Parameter | Estimate | Wald Z | Sig. | |
|---|---|---|---|---|
| Residual | 0.029149 | 2.236 | 0.025 | |
| Day [subject = id] | Variance | 0.000000a | . | . |
| Residual | 15.235632 | 2.449 | 0.014 | |
| Trial [subject = id] | Variance | 4.155172 | 0.000 | 1.000 |
a : this covariance parameter is redundant. The test statistic and confidence interval cannot be computed. Sig.: the p-value (associated with the correlation).
3.6. Practical Aspects
Enterococcal strains with a view to be used as protective or starter/adjunct cultures in biopreservation of foods, must usually be selected on the basis of the safety aspects which frequently are the absence of virulence and antibiotic resistance traits. Effectively, E. lactis 4CP3 strain was previously verified as nonhaemolytic, gelatinase negative, sensitive to vancomycin and other clinically relevant antibiotics and lacked known antibiotic resistance genes and several significant virulence factors [21]. Therefore, the presence of E. lactis 4CP3 in meat does not appear to represent a health risk.
4. Conclusion
To the best of our knowledge, this is the first report on the application of a multiple enterocin-producing E. lactis strain to control L. monocytogenes in artificially contaminated raw beef meat during refrigerated storage. Based on the obtained results, E. lactis 4CP3 strain might be useful as natural biopreservative against L. monocytogenes in meat products.
Acknowledgments
This work was supported by a grant from the Ministry of High Education, Tunisia. The authors thank Dr. Stefano Morandi (Institute of Sciences of Food Production, Italian National Research Council (CNR ISPA), Milan, Italy) for providing them with the non-bacteriocin-producing E. faecium VC185 strain and the RAPD-PCR analysis.
Conflicts of Interest
The authors have declared that there are no conflicts of interest.
References
- 1.Islam M. S., Husna A. A., Islam M. A., Khatun M. M. Prevalence of Listeria monocytogenes in beef, chevon and chicken in Bangladesh. American Journal of Food Science and Health. 2016;2(4):39–44. [Google Scholar]
- 2.Lennox J. A., Etta P. O., John G. E., Henshaw E. E. Prevalence of Listeria monocytogenes in fresh and raw fish, chicken and beef. Journal of Advances in Microbiology. 2017;3(4):1–7. [Google Scholar]
- 3.Ismaiel A. A.-R., Ali A. E.-S., Enan G. Incidence of Listeria in Egyptian meat and dairy samples. Food Science and Biotechnology. 2014;23(1):179–185. doi: 10.1007/s10068-014-0024-5. [DOI] [Google Scholar]
- 4.Abdellatif S. H., Abdel-Shafi S., Ali A. E., Ismaiel A. A. Inhibition of two Listeria strains in vitro and in situ by nisin, onion and garlic juices separately and in combinations. Wulfenia Journal. 2018;25(1):194–206. [Google Scholar]
- 5.Martín B., Perich A., Gómez D., et al. Diversity and distribution of Listeria monocytogenes in meat processing plants. Food Microbiology. 2014;44:119–127. doi: 10.1016/j.fm.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Dortu C., Huch M., Holzapfel W. H., Franz C. M. A. P., Thonart P. Anti-listerial activity of bacteriocin-producing Lactobacillus curvatus CWBI-B28 and Lactobacillus sakei CWBI-B1365 on raw beef and poultry meat. Letters in Applied Microbiology. 2008;47(6):581–586. doi: 10.1111/j.1472-765X.2008.02468.x. [DOI] [PubMed] [Google Scholar]
- 7.EFSA. The european union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA Journal. 2013;11(4):p. 3129. doi: 10.2903/j.efsa.2013.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson P. M., Cekmer H. B., Monu E. A., Techathuvanan C. The use of natural antimicrobials in food: an overview. Handbook of Natural Antimicrobials for Food Safety and Quality. 2015:1–27. [Google Scholar]
- 9.Widyastuti Y., Febrisiantosa A. The role of lactic acid bacteria in milk fermentation. Journal of Food and Nutrition Sciences. 2014;05(04):435–442. doi: 10.4236/fns.2014.54051. [DOI] [Google Scholar]
- 10.Pilet M.-F., Leroi F. Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food And Beverage Biopreservation. Cambridge, UK: Woodhead Publishing; 2011. Applications of protective cultures, bacteriocins and bacteriophages in fresh seafood and seafood products; pp. 324–347. [Google Scholar]
- 11.Melero B., Vinuesa R., Diez A. M., Jaime I., Rovira J. Application of protective cultures against Listeria monocytogenes and Campylobacter jejuni in chicken products packaged under modified atmosphere. Poultry Science. 2013;92(4):1108–1116. doi: 10.3382/ps.2012-02539. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Haliem M. E. F., Tartour E., Enan G. Characterization, production and partial purification of a bacteriocin produced by Lactobacillus plantarum LPS10 isolated from pickled lives. Research Journal of Pharmaceutical, Biological and Chemical Sciences. 2016;7(5):2362–2371. [Google Scholar]
- 13.Smaoui S., Elleuch L., Ben Salah R., et al. Efficient role of BacTN635 on the safety properties, sensory attributes, and texture profile of raw minced meat beef and chicken breast. Food Additives and Contaminants - Part A Chemistry, Analysis, Control, Exposure and Risk Assessment. 2014;31(2):218–225. doi: 10.1080/19440049.2013.873144. [DOI] [PubMed] [Google Scholar]
- 14.Smaoui S., Hsouna A. B., Lahmar A., et al. Bio-preservative effect of the essential oil of the endemic Mentha piperita used alone and in combination with BacTN635 in stored minced beef meat. Meat Science. 2016;117:196–204. doi: 10.1016/j.meatsci.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Paari A., Kanmani P., Satishkumar R., Yuvaraj N., Pattukumar V., Arul V. Potential function of a novel protective culture Enterococcus faecium-MC13 isolated from the gut of Mughil cephalus: safety assessment and its custom as biopreservative. Food Biotechnology. 2012;26(2):180–197. doi: 10.1080/08905436.2012.670891. [DOI] [Google Scholar]
- 16.Vandera E., Lianou A., Kakouri A., Feng J., Koukkou A.-I., Samelis J. Enhanced control of Listeria monocytogenes by Enterococcus faecium KE82, a multiple enterocin-producing strain, in different milk environments. Journal of Food Protection. 2017;80(1):74–85. doi: 10.4315/0362-028X.JFP-16-082. [DOI] [PubMed] [Google Scholar]
- 17.Aspri M., Field D., Cotter P. D., Ross P., Hill C., Papademas P. Application of bacteriocin-producing Enterococcus faecium isolated from donkey milk, in the bio-control of Listeria monocytogenes in fresh whey cheese. International Dairy Journal. 2017;73:1–9. doi: 10.1016/j.idairyj.2017.04.008. [DOI] [Google Scholar]
- 18.Chakchouk-Mtibaa A., Elleuch L., Smaoui S., et al. An antilisterial bacteriocin BacFL31 produced by Enterococcus faecium FL31 with a novel structure containing hydroxyproline residues. Anaerobe. 2014;27:1–6. doi: 10.1016/j.anaerobe.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Coelho M. C., Silva C. C. G., Ribeiro S. C., Dapkevicius M. L. N. E., Rosa H. J. D. Control of Listeria monocytogenes in fresh cheese using protective lactic acid bacteria. International Journal of Food Microbiology. 2014;191:53–59. doi: 10.1016/j.ijfoodmicro.2014.08.029. [DOI] [PubMed] [Google Scholar]
- 20.Weiss J., Zhong Q., Harte F., Davidson P. M. Micro- and nanoparticles for controlling microorganisms in foods. In: Pabst G., Kucerka N., Nieh M. P., Katsaras J., editors. Liposomes, Lipid Bilayers and Model Membranes: From Basic Research to Technology. Boca Raton, FL, USA: CRC Press; 2014. [Google Scholar]
- 21.Ben Braïek O., Cremonesi P., Morandi S., Smaoui S., Hani K., Ghrairi T. Safety characterisation and inhibition of fungi and bacteria by a novel multiple enterocin-producing Enterococcus lactis 4CP3 strain. Microbial Pathogenesis. 2018;118:32–38. doi: 10.1016/j.micpath.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Morandi S., Silvetti T., Miranda Lopez J. M., Brasca M. Antimicrobial activity, antibiotic resistance and the safety of lactic acid bacteria in raw milk valtellina casera cheese. Journal of Food Safety. 2015;35(2):193–205. doi: 10.1111/jfs.12171. [DOI] [Google Scholar]
- 23.French Standardization Association (AFNOR) NF EN ISO. 11290. Plaine Saint-Denis, France: 2005. Microbiology of food and animal feeding stuffs: Horizontal method for the detection and enumeration of Listeria monocytogenes-Part 2: Enumeration method. [Google Scholar]
- 24.Ghrairi T., Frere J., Berjeaud J. M., Manai M. Purification and characterization of bacteriocins produced by Enterococcus faecium from Tunisian rigouta cheese. Food Control. 2008;19(2):162–169. doi: 10.1016/j.foodcont.2007.03.003. [DOI] [Google Scholar]
- 25.Cremonesi P., Castiglioni B., Malferrari G., et al. Technical note: Improved method for rapid DNA extraction of mastitis pathogens directly from milk. Journal of Dairy Science. 2006;89(1):163–169. doi: 10.3168/jds.S0022-0302(06)72080-X. [DOI] [PubMed] [Google Scholar]
- 26.Andrighetto C., Zampese L., Lombardi A. RAPD-PCR characterization of lactobacilli isolated from artisanal meat plants and traditional fermented sausages of Veneto region (Italy) Letters in Applied Microbiology. 2001;33(1):26–30. doi: 10.1046/j.1472-765X.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- 27.Ben Braïek O., Ghomrassi H., Cremonesi P., et al. Isolation and characterisation of an enterocin P-producing Enterococcus lactis strain from a fresh shrimp (Penaeus vannamei) Antonie van Leeuwenhoek Journal of Microbiology. 2017;110(6):771–786. doi: 10.1007/s10482-017-0847-1. [DOI] [PubMed] [Google Scholar]
- 28.El Abed N., Kaabi B., Smaali M. I., et al. Chemical composition, antioxidant and antimicrobial activities of thymus capitata essential oil with its preservative effect against Listeria monocytogenes inoculated in minced beef meat. Evidence-Based Complementary and Alternative Medicine. 2014;2014:11. doi: 10.1155/2014/152487.152487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.International Organization for Standardization (ISO) Microbiology of food and animal feeding stuffs—Horizontal method for the enumeration of psychrotrophic microorganisms. 2001;(17410)
- 30.John U. V., Carvalho J. Enterococcus: Review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Frontiers in Biology. 2011;6(5):357–366. doi: 10.1007/s11515-011-1167-x. [DOI] [Google Scholar]
- 31.Kennedy J., Jackson V., Blair I. S., McDowell D. A., Cowan C., Bolton D. J. Food safety knowledge of consumers and the microbiological and temperature status of their refrigerators. Journal of Food Protection. 2005;68(7):1421–1430. doi: 10.4315/0362-028X-68.7.1421. [DOI] [PubMed] [Google Scholar]
- 32.Morandi S., Silvetti T., Brasca M. Biotechnological and safety characterization of Enterococcus lactis, a recently described species of dairy origin. Antonie van Leeuwenhoek-Journal of Microbiology. 2013;103(1):239–249. doi: 10.1007/s10482-012-9806-z. [DOI] [PubMed] [Google Scholar]
- 33.Rehaiem A., Belgacem Z. B., Edalatian M. R., et al. Assessment of potential probiotic properties and multiple bacteriocin encoding-genes of the technological performing strain Enterococcus faecium MMRA. Food Control. 2014;37(1):343–350. doi: 10.1016/j.foodcont.2013.09.044. [DOI] [Google Scholar]
- 34.RASFF. The Rapid Alert System for Food and Feed: Annual Report. Luxembourg, Luxembourg: Publications Office of the European Union; 2017. [Google Scholar]
- 35.Pragalaki T., Bloukas J. G., Kotzekidou P. Inhibition of Listeria monocytogenes and Escherichia coli O157:H7 in liquid broth medium and during processing of fermented sausage using autochthonous starter cultures. Meat Science. 2013;95(3):458–464. doi: 10.1016/j.meatsci.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 36.Giello M., La Storia A., De Filippis F., Ercolini D., Villani F. Impact of Lactobacillus curvatus 54M16 on microbiota composition and growth of Listeria monocytogenes in fermented sausages. Food Microbiology. 2018;72:1–15. doi: 10.1016/j.fm.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Rivas F. P., Castro M. P., Vallejo M., Marguet E., Campos C. A. Sakacin Q produced by Lactobacillus curvatus ACU-1: functionality characterization and antilisterial activity on cooked meat surface. Meat Science. 2014;97(4):475–479. doi: 10.1016/j.meatsci.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 38.de Souza Barbosa M., Todorov S. D., Ivanova I., Chobert J.-M., Haertlé T., de Melo Franco B. D. G. Improving safety of salami by application of bacteriocins produced by an autochthonous Lactobacillus curvatus isolate. Food Microbiology. 2015;46:254–262. doi: 10.1016/j.fm.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Woraprayote W., Malila Y., Sorapukdee S., Swetwiwathana A., Benjakul S., Visessanguan W. Bacteriocins from lactic acid bacteria and their applications in meat and meat products. Meat Science. 2016;120:118–132. doi: 10.1016/j.meatsci.2016.04.004. [DOI] [PubMed] [Google Scholar]