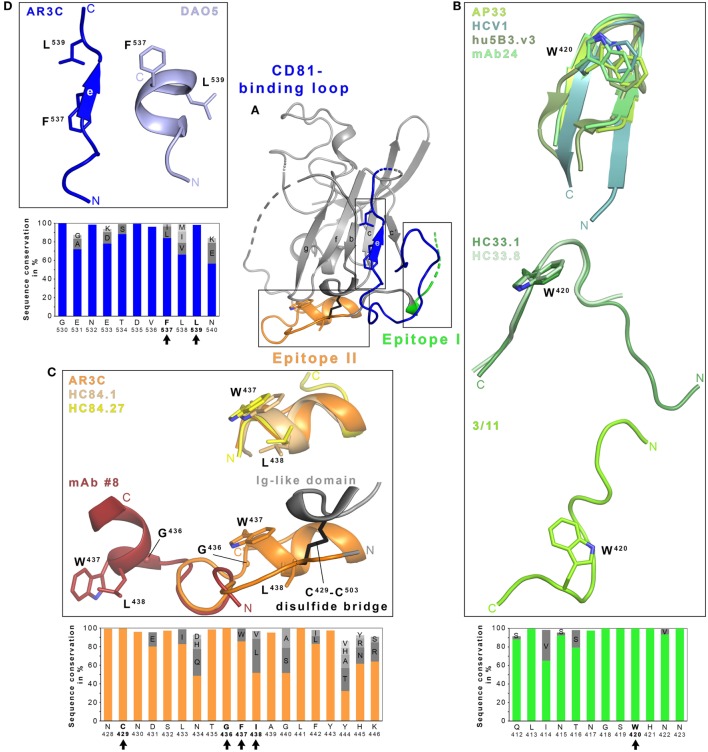
Figure 1.
Structural flexibility of the hepatitis C virus (HCV) E2 glycoprotein. (A) Cartoon representation of the E2 ectodomain crystallized in complex with AR3C Fab (PDB 4MWF). The composite CD81-binding site consisting of epitope I (aa412–423; green), epitope II (aa428–446; orange), and the CD81-binding loop (aa518–542; blue) is highlighted in color and sidechains of selected residues are displayed as sticks. (B–D) Close-up views of the three antigenic sites mentioned above. (B) Epitope I is disordered in the context of the E2 structure but a synthetic epitope peptide folds as β-hairpin in complex with neutralizing antibodies (nAbs) AP33 (PDB 4GAJ), HCV1 (PDB 4DGV), hu5B3.v3 (PDB 4HS8), and mAb24 (PDB 5VXR) (upper panel). By contrast, the same peptide adopts two distinct extended conformations in complex with nAbs HC33.1 (PDB 4XVJ) and HC33.8 (PDB 5FGC) (middle panel) or 3/11 (PDB 4WHY; bottom panel), respectively. The peptide in the nAb HC33.4 complex (PDB 5FGB) adopts an amino acid backbone conformation identical to the one in complex with nAb HC33.8 and is not shown for simplicity. (C) Superposition of the epitope II peptide structure in complex with HC84.1 and HC84.27 Fabs (PDB 4JZN and 4JZO, respectively) onto the E2 ectodomain structure (upper panel) reveals a conserved 1.5-turn α-helix (aa437–442) with an extended C-terminal segment containing aa443–446. Superposition of the N-terminal loop of epitope II (aa430–434) from the peptide structure in complex with mAb #8 (PDB 4HZL) onto its counterparts in the E2 structure suggests that the short α-helix flips out to expose residues W437 and L438 for mAb #8 binding (bottom panel). (D) Residues 532–540 of the CD81-binding loop were observed in an extended conformation in the context of the E2 ectodomain structure (A) and in a helical conformation in the DAO5 Fab–E2 peptide complex structure (PDB 5NPJ) suggesting thereby a putative open and closed conformation of the immunoglobulin-like domain. Amino acid sequence conservation of the respective antigenic site was calculated across the six HCV genotypes for 481 isolate sequences (100 sequences each for genotypes 1, 2, 3, 6, and 70, 9 and 2 sequences for genotypes 4, 5, and 7, respectively) obtained and analyzed from the ViPR database (http://www.viprbrc.org) and is shown below each close-up view. Residues with side chains shown as sticks are highlighted by black arrows.