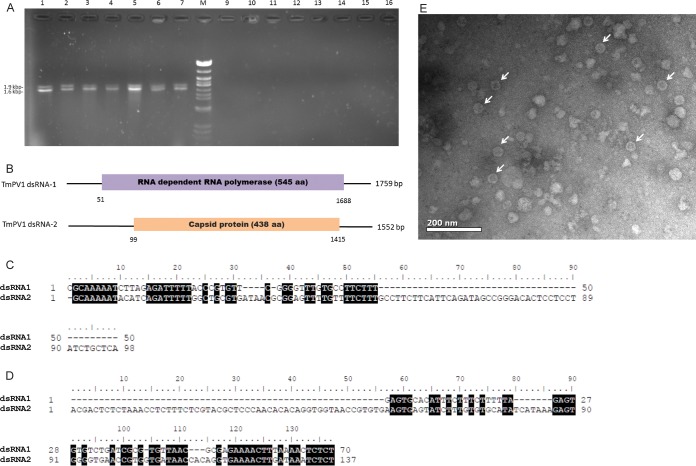
FIG 1 .
(A) dsRNA extracted from Talaromyces marneffei. (B) Schematic representation of the genome organization of TmPV1 dsRNA-1 and dsRNA-2. (C and D) Conserved 5′-UTR (C) and 3′-UTR (D) sequence elements in TmPV1 dsRNA-1 (top rows) and dsRNA-2 (bottom rows). (E) Transmission electron microscopy of TmPV1 purified from T. marneffei isolate PM40. (A) Lanes 1 to 7, two dsRNA bands were observed in 7 T. marneffei isolates; lanes 9 to 16, no dsRNA bands were observed in the remaining 48 T. marneffei isolates (7 selected isolates are shown); lane M, lambda DNA digested by Eco471 (AvaII). Sizes are indicated as kilobase pairs (left). (B) Open bars represent open reading frames (ORFs) encoding the putative RNA-dependent RNA polymerase or capsid protein. The 5′ and 3′ untranslated regions are indicated as single lines. The nucleotide positions of the initiation and termination codons are indicated below the border of the ORF. Nucleotides in the 5′-UTR (C) and 3′-UTR (D) of dsRNA-1 and dsRNA-2 that are identical are indicated with reverse highlighting. (E) Isometric, nonenveloped viral particles of 30 to 45 nm in diameter, compatible with partitiviruses, are indicated by white arrows. Bar = 200 nm.