Abstract
New techniques for single-cell analysis enable new discoveries in gene expression and systems biology. Time-dependent measurements on individual cells are necessary, yet the common single-cell analysis techniques used today require lysing the cell, suspending the cell, or long incubation times for transfection, thereby interfering with the ability to track an individual cell over time. Here a method for detecting mRNA expression in live single cells using molecular beacons that are transfected into single cells by means of nanofountain probe electroporation (NFP-E) is presented. Molecular beacons are oligonucleotides that emit fluorescence upon binding to an mRNA target, rendering them useful for spatial and temporal studies of live cells. The NFP-E is used to transfect a DNA-based beacon that detects glyceraldehyde 3-phosphate dehydrogenase and an RNA-based beacon that detects a sequence cloned in the green fluorescence protein mRNA. It is shown that imaging analysis of transfection and mRNA detection can be performed within seconds after electroporation and without disturbing adhered cells. In addition, it is shown that time-dependent detection of mRNA expression is feasible by transfecting the same single cell at different time points. This technique will be particularly useful for studies of cell differentiation, where several measurements of mRNA expression are required over time.
1. Introduction
Cell function, phenotype, and cycle state are dictated by expression and processing of RNA molecules. The advent of molecular biology techniques such as polymerase chain reaction (PCR) and microarrays made it possible to determine the phenotype of a cell population by measuring mRNA expression. However, it has been shown that daughter cells plated in the same environment may diverge phenotypically because gene expression is noisy within each single cell, [1–5] thus the results obtained from sampling a population of cells may mask what occurs at the single-cell level. New techniques such as quantitative PCR can be performed on single cells, [6] but require lysis of the cells, which prohibits the ability to collect spatial information and limits the temporal data obtained. Currently, resolving mRNA expression spatially in the cytoplasm of cells can be accomplished with fluorescent in situ hybridization (FISH), but requires fixing cells, limiting studies to discrete time points and preventing a dynamic study of the cell. [7]
To study mRNA expression in live cells, the most widely used probe is the molecular beacon (MB). Using MBs may facilitate the study of both spatial and temporal localization of mRNAs. These molecules consist of DNA or RNA oligonucleotides (native or modified) labeled at one end with a fluorescent tag and at the other with a quencher molecule. [8–13] The MB folds into a hairpin-like structure, positioning the fluorophore and quencher together such that fluorescence light emission is inhibited. The loop of the hairpin is an oligonucleotide sequence that is complementary to the target mRNA sequence, and upon binding, the MB hairpin opens and fluorescent light is emitted. This design achieves a high signal-to-noise ratio due to the up to 200-fold increase in fluorescence upon MB binding to the target mRNA. [9,14,15] A feature that is vital for mRNA studies in live single cells. [11,16–18] For example, Rhee et al. have shown that MBs can detect Oct-4 expression from a mixed population of stem cells as a method for stem cell detection and isolation using phenotype markers inside the cell. [11] Recently, Desai et al. used three MBs to detect alkaline phophatase, type I collagen, and osteocalcin mRNAs to follow the timing of differentiation from single-cell adipose stem cells into osteocytes. [16] Unique variants of MBs, e.g., ratiometric bimolecular beacons (RBMBs) have also been developed to further improve the signal-to-background of measurements of RNA expression in living cells. [19–21]
The addition of MBs that bind to mRNA in the cytoplasm and generate temporary double-stranded RNA is similar to RNA silencing processes inherent to the cell. However, the complexity and length of the double-stranded structures of microRNA and siRNA precursors [22,23] vastly differ from the structure of MBs, making it unlikely that MBs will induce mRNA degradation. Indeed, studies using MBs to target the mRNA of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and K-ras (an enzyme involved in receptor activation) [18] and Oct-4 mRNA [11] have shown that mRNA levels are not significantly altered by the presence of the MBs. Furthermore, MBs are not expected to interfere with translation of the target mRNA into protein. [24] These studies indicate that MBs do not have a deleterious effect to living cells and can be used to monitor gene expression in single cells without altering cell function.
MBs have been delivered into live cells using bulk transfection methods such as streptolysin-O (SLO), [18] gold nanoprobes, [25] microporation, [13,26] cell penetrating peptide, [27] and standard bulk electroporation. [28] However, single-cell transfection of MBs has only been accomplished by microinjection, [12,19] a technique that is technically demanding and relatively invasive, particularly for experiments where multiple transfections of the same cell are warranted. Each of these methods can induce significant cellular stress, which can potentially affect their phenotype and limit their usefulness for time-dependent studies. [24,25] Methods that require cells in suspension, also suffer the disadvantage that cells must be detached from the culture dish using enzymes, for the transfection procedure, and then re-plated for image analysis. Therefore, these methods cannot be used to either track the same single cell over time or image the cells immediately after transfection.
Despite the variety of transfection techniques available to deliver MBs, the only minimally invasive technique that would enable transfection of single cells within a population and immediate optical/fluorescent imaging after transfection is nanofountain probe electroporation (NFP-E). [29,30] The NFP-E system consists of a microfluidic device with a nanoscale cantilever probe tip together with an integrated electronic component and software to deliver a localized voltage to a cell membrane for electroporation. The system can be coupled with standard equipment used for cell research such as an inverted microscope, petri dishes for cell culture, and micromanipulator. Recently, we have reported NFP electroporation of single cells to efficiently transfect various types of molecules while maintaining high cell viability. [29] Here, we present single-cell transfection of DNA- and RNA-based MBs [18–20] using the NFP-E system to demonstrate the potential for this technology to enable studies of gene expression and cell phenotype at a single-cell level. In addition, we demonstrate that the NFP-E system can be used to repeatedly transfect the same cell over a period of 24 h as a proof of concept for time-dependent detection of different molecules, which would be particularly useful for monitoring cell differentiation using MBs.
2. Results and Discussion
2.1. NFP-E System for Electroporation of MBs
An NFP chip containing a 1D array of 12 probes was packaged such that fluidic access and a Ag wire electrode are directly connected to the microchip reservoir. [30] To reduce the volume of solution containing MBs needed for this work, the NFP chip and packaging were modified from the previously published design containing an array of probes [29,30] to generate a one-probe NFP-E unit requiring only 1 μL of solution per experiment ( Figure 1a). We were able to reduce the volume to only 1 μL by switching the electrode from the back of the packaging to the front directly above the on-chip reservoir. With this modification, the electrode is always in contact with the electroporation solution, even when using small volumes of solution. Furthermore, the modified packaging makes it possible to recover unused solution such that several experiments transfecting at least 50 cells each can be performed with 1 μL of transfecting solution. The new packaging design did not require changing the electroporation protocol; therefore, we expect the mechanism of NFP electroporation with this packaging to be identical to that previously published.
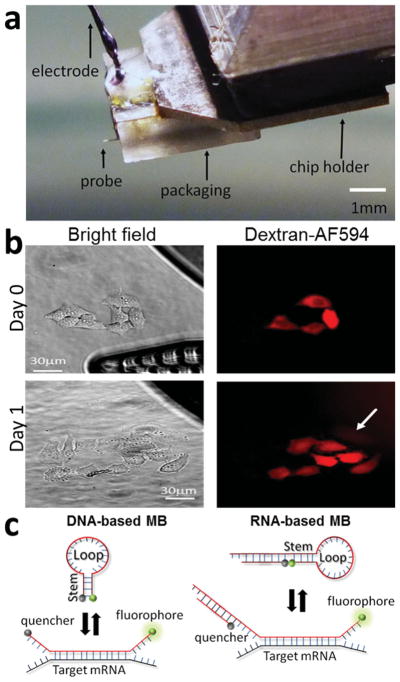
Figure 1.
a) Image of the NFP chip with only one probe and polycarbonate packaging unit that incorporates a microchannel for loading the NFP chip and an electrode to apply voltage.[29] b) Four HeLa cells transfected on day 0 with Dextran-AF594 and imaged after a 24 h incubation (day 1) showing that the electroporated cells divided. The NFP-E probe is shown on the bright field day 0 picture. c) Schematic structures of the two types of MBs used in this study showing how fluorescence is induced when the MB binds to its mRNA target.[18,19]
In prior work we demonstrated that NFP electroporation is gentle to cells and that more than 90% of electroporated cells are viable 4 h after transfection. [29] However, to follow gene expression and cell differentiation, cells need to stay healthy for several hours or days after electroporation. To further explore cell health after NFP electroporation, we transfected single HeLa cells with dextran tagged with Alexa fluor-594 (Dextran-AF594) using the NFP-E and determined their state after 24 h of incubation (Figure 1b). A representative experiment in Figure 1b shows electroporated cells remain healthy and undergo cell division, correlating with the previously published high viability observed 4 h post transfection. [26] These results indicate that the NFP-E system can be used to follow the same single cells over several hours and could be a useful tool for transfecting different molecules or stimuli over time.
In this study, we tested two different MB designs, one DNA-based and the other RNA-based, that target two different mRNAs (Figure 1c). The DNA-based MB (DMB) contains an unmodified DNA backbone that efficiently targets the mRNA of the GAPDH enzyme. [18] The GAPDH enzyme performs an important role in the glycolysis pathway of a cell, so a healthy cell should express GAPDH mRNA constantly throughout the cell cycle. [31] This DMB contains a fluorophore (carboxyfluorescein, FAM) at the 5′ end and a quencher (BHQ1) at the 3′ end. When not in the presence of its mRNA GAPDH target, it folds in a hairpin configuration. Upon binding to its target, the hairpin opens and the FAM molecule emits fluorescence (Figure 1c). Similarly, the RNA-based MB (RMB) contains a Cy5 fluorophore on the 5′ end and an Iowa Black RQ-Sp quencher at the 3′ end. The backbone consists of two 2′-O-methyl-modified RNA molecules hybridized together [19,20] (Figure 1c). One of the RNA oligonucleotides is longer than the other one, allowing its 5′ end to fold on itself to form a hairpin structure. The double stranded sequence of the RMB contains a 5′ UU overhang, facilitating nuclear export in live cells. [19] This RMB construct has the potential to contain a second fluorophore to monitor delivery into the cell that could be used to normalize the signal and quantify gene expression. [19] For this study, however, we did not employ the RMB with a second fluorophore (Figure 1c).
2.2. Transfection of the DNA-Based MB Using the NFP-E System
We first transfected HeLa cells with the DMB using the NFP-E system. Because GAPDH is distributed throughout the cytoplasm, the DMB fluoresced throughout the cell immediately after transfection (Figure 2a). Ten minutes post transfection, the fluorescent signal indicated that the DMB had localized inside the nucleus (Figure 2a). Nuclear import of the GAPDH-DMB was observed after a few minutes regardless of where the probe tip was positioned on the cell (not shown). These results correlate with other studies showing that small unmodified oligonucleotides, such as the DMB, are sequestered inside the nucleus due to active nuclear import or binding to nuclear proteins. [24,32,33] While this localization is detrimental to the utility of unmodified DNA-based MBs for monitoring gene expression when other transfection methods are used, NFP electroporation enables real time signal acquisition, allowing the observation of the GAPDH-DMB signal before transport into the nucleus occurs. Indeed, by electroporating a cell at one of its extremes (white arrow in Figure 2b) and simultaneously imaging the fluorescence signal, we were able to observe a fluorescent front diffusing away from the electroporation spot (Figure 2b, 30 s). After 4 min, the fluorescent signal was distributed throughout the entire cytoplasm (Figure 2b). This ability to spatially control the transfection site and visualize how the transfected molecule diffuses or is transported inside the cell is a useful feature of NFP electroporation, particularly for gene therapy studies where researchers are interested in determining the fate of injected or transfected genes within a cell. [34] Currently, to determine the rate of diffusion of DNA nucleotides inside cells, researchers use microinjection to flood a cell with a fluorescently tagged DNA molecule and then photo-bleach one spot inside the cell using a laser. [32] This technique actually measures the intrinsic motion of DNA molecules inside a cell, rather than the true diffusion. By contrast, the NFP-E technique enables the measurement of gradients and moving concentration fronts in real time.
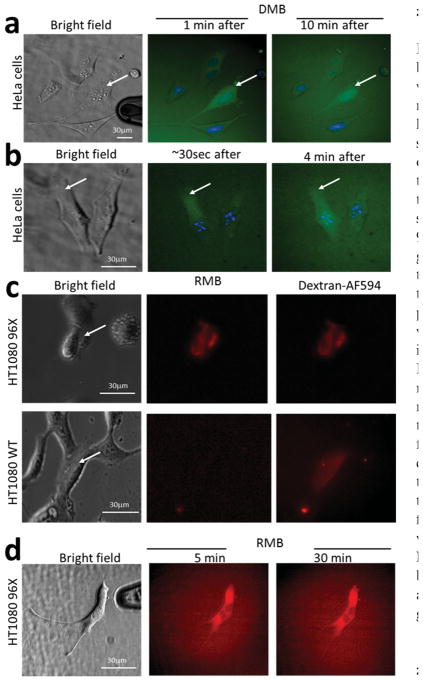
Figure 2.
Transfection of DNA-based (DMB) and RNA-based (RMB) molecular beacons by NFP electroporation. a,b) HeLa cells with Hoescht-stained nuclei (blue) were transfected with a DMB specific for GAPDH mRNA that fluoresces green upon binding.[18] a) After electroporation, green fluorescence was immediately detected with the majority of DMB transported inside the nucleus within 10 min. b) Diffusion of the DMB was observed in real time by monitoring the fluorescence signal front through the cytoplasm. c,d) HT1080 cells transfected with the RMB tagged with a Cy5 fluorophore specific for a nucleotide sequence engineered in the untranslated region of a GFP mRNA.[19] HT1080 96X cells constitutively express 96 tandem repeats of this sequence for signal enhancement. As a positive control for electroporation, dextran-AF594 was transfected in tandem with the RMB. HT1080 WT cells did not show any RMB signal after transfection. d) Fluorescence images of the RMB transfected into HT1080–96X cells 5 min and 30 min after electroporation show that the RMB did not localize in the nucleus after transfection.
2.3. Transfection of RNA-Based MB Using the NFP-E System
Because small oligonucleotides, with unmodified DNA backbones, localize in the nucleus quickly after electroporation, we tested other types of MBs. The RMB molecule is a RNA molecule that contains 2′-O-methyl modifications, making it less sensitive to nucleases (RNAses). Moreover, the double stranded nature of the RMB (Figure 1c) and the 3′-UU overhang resemble structural features of siRNA that facilitate transport out of the nucleus, [35] inhibiting its accumulation in this organelle. [19,20] The RMB in this study targets a short nucleotide sequence, engineered to repeat in tandem 96 times in the 5′-untranslated region of a mRNA encoding green fluorescent protein (GFP), and emits signal only when this construct is present. [19] Using NFP electroporation, we transfected the RMB together with dextran-AF594 (as a positive control of electroporation) into single HT1080 cells, which stably express this construct (HT1080 96X), [36] and into wild type HT1080 (HT1080 WT) cells (Figure 2c). In the HT1080 96X cells, we observed immediately after electroporation red fluorescence indicating RMB binding to its target mRNA. No fluorescence from the RMB was observed in the transfected HT1080 WT despite the presence of fluorescence from dextran-AF594, which indicated the occurrance of succcessful transfection. Contrary to the transfection of the DMB, the RMB did not localize in the nucleus within the 30 min that we monitored the transfected cells (Figure 2d). This confirms the findings reported by Chen et al., [19] where an RMB was transfected using a microporation technique. Thus, using NFP electroporation to transfect single cells with RNA-based MBs, like the RMB used in this study, would provide a method to detect over time the expression of endogenous genes of interest in individual cells within a population.
2.4. Transfection into the Same Single Cell Over Time
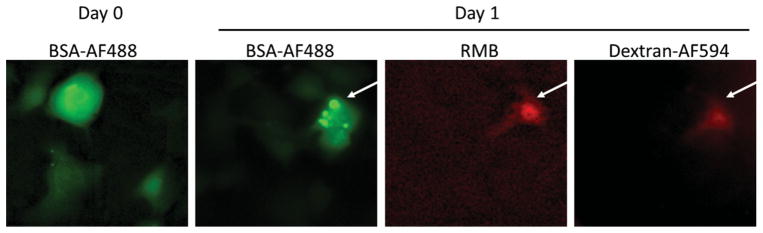
Time-dependent monitoring of several mRNAs in single cells can be accomplished using multiple MBs to detect different genes. [37] However, delivery of the MBs is currently only possible using invasive techniques like microinjection or nanoneedles. [38] The stress to the cell caused by mechanically puncturing the membrane can cause significant damage to cell architecture, and possibly function, [36] thereby preventing the delivery of probes multiple times over the course of a long-term study. We hypothesize that NFP electroporation could also be used for long term, time-dependent single-cell studies, and may be superior to other techniques by causing less stress on the cell due to the localized nature of the electric field. [29] As a proof of concept, we transfected single cells on one day (day 0) with bovine serum albumin tagged with Alexa Fluor 488 (BSA-AF488) and then again with a solution of RMB and dextran-AF594 the next day (Day 1). Specifically, we transfected a group of HT1080–96X cells with BSA-AF488 (Figure 3, Day 0-BSA-AF488) and incubated the plate overnight. The following day, we located the transfected cells using the fluorescence from the BSA-AF488 and chose one cell to transfect using the RMB and dextran-AF594 solution (Figure 3, Day 1). On day 1 some of the transfected BSA-AF488 in the target cell had accumulated into certain areas of the cell, likely vacuoles. After electroporation to transfect RMB/dextran-AF594, we observed that these new molecules diffused freely inside the cell. The experimental data are a proof-of-concept for the potential of NFP electroporation for multiple administrations of MBs into the same cell.
Figure 3.
Transfection of the same cells over time (~24 h) by NFP electroporation. A group of HT1080–96X cells were transfected on day 0 with BSA-AF488 and incubated overnight. The following day, Day 1, the transfected group of cells showed that BSA-AF488 localized within an organelle of the cell. In addition, on day 1, one cell from this group (white arrow) was transfected again with a solution of RMB and dextran-AF594 (positive control).
3. Conclusion
We demonstrated that NFP electroporation can be used to deliver MBs into single cells for real-time imaging and analysis of gene expression. We efficiently transfected two different MB probes that have been previously used to detect mRNA in live cells. We also presented two proof of concept experiments for time-dependent studies: (a) showing that transfected HeLa cells were healthy and had divided within 24 h post transfection, and (b) transfecting the same single cells with different color fluorophores on two sequential days. Together, these preliminary experimental results indicate that NFP electroporation does not significantly stress the cell or interrupt the cell cycle; a more extensive study to collect statistics on various cell types and protocols is needed to fully assess the potential of the approach. Hence, by using NFP electroporation to transfect RNA-based MBs into single cells, the technique offers the potential to monitor mRNAs expression over time in the same cell. This capability could be used to monitor gene expression in single cells during somatic cell reprogramming (e.g., detection of pluripotency markers) and stem cell differentiation, potentially accelerating the progress of research in embryonic and induced pluripotent stem cells (iPSC). Likewise, single-cell gene expression can be employed in the identification of regulatory networks (e.g., metabolomics) and mechanism of disease in iPSC-derived cells.
4. Experimental Section
Molecular Beacons
The MB targeting GAPDH was produced using standard DNA synthesis. This DMB is comprised of the following nucleotide sequence and attached molecules: 5′-FAMCGACGGAGTCCTTCCACGATACCACGTCG- BHQ1–3′. The RMB targeting mRNA from GFP was prepared as previously described and is composed of two hybridized 2-O′-methyl-modified RNA oligonucleotides. [13] The sequence of the longer oligonucleotide was: 5′CF640R- mCmUmUmC mGmUmC mCmAmC mAmAmA mCmAmC mAmAmC mUmCmC mU mGmAmAmG mGmAmC mGmGmC mAmGmC mGmUmG mCmAmG mCmUmC mUmU-3′. The self-complementary domains that promote the formation of the hairpin are underlined. The shorter oligonucleotide contains the following sequence: 5′-mGmAmG mCmUmG mCmAmC mGmCmU mGmCmC mGmUmC-IBRS-3′. The 3′ end of this oligonucleotide contains an Iowa Black RQ-Sp quencher (Integrated DNA technologies, IDT) that quenches the signal emitted by CF640R when there is no target mRNA present.
Cell Culture for Passage and Transfection
HeLa cells were purchased from ATCC (Manassas, VA). Engineered HT1080 (HT1080- GFP-96mer and HT1080-GFP) as well as wild type HT1080 were provided by the Tsourkus laboratory. [20,21] Cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies), which contained: DMEM with phenol red supplemented with 10% fetal bovine serum (FBS, Life Technologies) and 1% Pen/ Strep (Life Technologies), and were incubated at 37 °C with 5% CO2. Engineered HT1080 cells were cultured in complete DMEM with 10 μg mL−1 blasticidin.
To prepare cells for transfection experiments, we plated HeLa and HT1080 cells in 2 cm-diameter glass bottom petri dishes with complete DMEM supplemented with 20% FBS instead of 10% one day before the experiment. The glass area in the petri dish contained approximately ≈50 000 cells with 800 μL of complete media. The day of the experiment, the complete DMEM media was replaced with DMEM containing Hepes (Life Technologies) and no Phenol Red. For experiments that required maintaining the cells live for a second round of electroporation, the DMEM-Hepes media was removed once the first electroporation was over and new complete DMEM with phenol Red was added. The cells were placed in the incubator at 37 °C with 5% CO2 until needed.
Electroporation of DMB and RMB Using the NFP-E System
For electroporation of the DMB, a solution of the MB in 1× PBS was prepared with a final concentration of 50 × 10−6 M. For electroporation of the RMB, the molecule was added to a solution containing 1× PBS and dextran-Alexa Fluor 594 (Life Technologies), adjusting the final concentration of RMB to 5 × 10 −6 M. 1 μL of the solution was then loaded into the NFP packaging by syringe and applied pressure using a FemtoJet (Eppendorf) pump. An InjectMan (Eppendorf) manipulator mounted on the microscope stage to position the NFP tip in contact with the target cells was used. The same electroporation parameters of three pulses of 0.5 s, 20 V at 200 Hz were applied to both HeLa and HT1080 cells upon contact of the cantilever tip with the target cell.
Image Acquisition
Experiments were performed using an Eclipse Ti Nikon inverted microscope with motorized stage and automatic objective changer coupled to a CCD Cool SNAP HQ2 camera (Photometrics). Fluorescent light was provided by a Nikon Intesilight C-HGFIE. A super plan fluor ELWD 40× Ph2 objective was used to aquire bright and fluorescent light images. The images were aquired and processed with Nikon Elements software.
Acknowledgments
This work was supported in part by a National Science Foundation grant [IIP-1330151] awarded to iNfinitesimal LLC, and grant [IIP-114562] awarded to H.D.E.; the National Institutes of Health award [1R41GM101833–01] to H.D.E.; a Career Award [0953583] awarded to A.T.; and Nanomedicine Development Center award [PN2EY018244] to G.B.
Contributor Information
Dr. Juan P. Giraldo-Vela, iNfinitesimal LLC, Skokie, IL 60077, USA. Department of Mechanical Engineering, Northwestern University, Evanston, IL 60208, USA
Dr. Wonmo Kang, iNfinitesimal LLC, Skokie, IL 60077, USA. Department of Mechanical Engineering, Northwestern University, Evanston, IL 60208, USA
Dr. Rebecca L. McNaughton, iNfinitesimal LLC, Skokie, IL 60077, USA. Department of Mechanical Engineering, Northwestern University, Evanston, IL 60208, USA
Dr. Xuemei Zhang, Department of Bioengineering, University of Pennsylvania, Philadelphia, PA 19104, USA
Brian M. Wile, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA 30332, USA
Prof. Andrew Tsourkas, Department of Bioengineering, University of Pennsylvania, Philadelphia, PA 19104, USA
Prof. Gang Bao, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA 30332, USA
Prof. Horacio D. Espinosa, iNfinitesimal LLC, Skokie, IL 60077, USA. Department of Mechanical Engineering, Northwestern University, Evanston, IL 60208, USA
References
- 1.Kalisky T, Blainey P, Quake SR. Annu Rev Genet. 2011;45:431. doi: 10.1146/annurev-genet-102209-163607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulsson J. Nature. 2004;427:415. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 3.Balazsi G, van Oudenaarden A, Collins JJ. Cell. 2011;144:910. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 5.Raj A, van Oudenaarden A. Cell. 2008;135:216. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong JF, Chen Y, Marcus JS, Scherer A, Quake SR, Taylor CR, Weiner LP. Lab Chip. 2008;8:68. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Femino A, Fay FS, Fogarty K, Singer RH. Science. 1998;280:585. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 8.Monroy-Contreras R, Vaca L. J Nucl Acids. 2011;2011:741723. doi: 10.4061/2011/741723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao G, Rhee WJ, Tsourkas A. Annu Rev Biomed Eng. 2009;11:25. doi: 10.1146/annurev-bioeng-061008-124920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen AK, Rhee WJ, Bao G, Tsourkas A. Methods Mol Biol. 2011;714:159. doi: 10.1007/978-1-61779-005-8_10. [DOI] [PubMed] [Google Scholar]
- 11.Rhee WJ, Bao G. BMC Biotechnol. 2009;9:30. doi: 10.1186/1472-6750-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsourkas A, Bao G. Brief Funct Genom Proteomics. 2003;1:372. doi: 10.1093/bfgp/1.4.372. [DOI] [PubMed] [Google Scholar]
- 13.Tsourkas A, Behlke MA, Bao G. Nucl Acids Res. 2002;30:5168. [PMC free article] [PubMed] [Google Scholar]
- 14.Tyagi S, Kramer FR. Nat Biotechnol. 1996;14:303. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi S, Bratu DP, Kramer FR. Nat Biotechnol. 1998;16:49. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 16.Desai HV, Voruganti IS, Jayasuriya C, Chen Q, Darling EM. Tissue Eng Part A. 2013;19:40. doi: 10.1089/ten.tea.2012.0127. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. Nucl Acids Res. 2005;33:1902. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nitin N, Rhee WJ, Bao G. Nucl Acids Res. 2009;37:4977. doi: 10.1093/nar/gkp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen AK, Davydenko O, Behlke MA, Tsourkas A. Nucl Acids Res. 2010;38:e148. doi: 10.1093/nar/gkq436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Song Y, Shah AY, Lekova V, Raj A, Huang L, Behlke MA, Tsourkas A. Nucl Acids Res. 2013;41:e152. doi: 10.1093/nar/gkt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang XM, Zajac AL, Huang LY, Behlke MA, Tsourkas A. PloS One. 2014;9:e85813. doi: 10.1371/journal.pone.0085813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ. J Biol Chem. 2004;279:42230. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 23.Khvorova A, Reynolds A, Jayasena SD. Cell. 2003;115:209. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 24.Tyagi S, Alsmadi O. Biophys J. 2004;87:4153. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan W, Zhang TT, Yang HJ, Diao W, Li N, Tang B. Anal Chem. 2013;85:10581. doi: 10.1021/ac402700s. [DOI] [PubMed] [Google Scholar]
- 26.Chen AK, Behlke MA, Tsourkas A. Nucl Acids Res. 2008;36:e69. doi: 10.1093/nar/gkn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nitin N, Santangelo PJ, Kim G, Nie SM, Bao G. Nucl Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desai HV, Voruganti IS, Jayasuriya C, Chen Q, Darling EM. Tissue Eng Part A. 2013;19:40. doi: 10.1089/ten.tea.2012.0127. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Kang WM, Yavari F, Jolandan MM, Vela GJP, Safi A, McNaughton RL, Parpoil V, Espinosa HD. Nano Lett. 2013;13:2448. doi: 10.1021/nl400423c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang W, McNaughton RL, Yavari F, Jolandan MM, Safi A, Espinosa HD. J Lab Autom. 2014;19:100. doi: 10.1177/2211068213495395. [DOI] [PubMed] [Google Scholar]
- 31.Barber RD, Harmer DW, Coleman RA, Clark BJ. Physiol Genom. 2005;21:389. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- 32.Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. J Biol Chem. 2000;275:1625. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- 33.Chen AK, Behlke MA, Tsourkas A. Nucl Acids Res. 2007;35:e105. doi: 10.1093/nar/gkm593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasiorowski JZ, Dean DA. Mol Ther. 2005;12:460. doi: 10.1016/j.ymthe.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. Nucl Acids Res. 2006;34:1369. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Song Y, Shah A, Lekova V, Raj A, Huang L, Behlke MA. Nucl Acids Res. 2013;41:e152. doi: 10.1093/nar/gkt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medley CD, Drake TJ, Tomasini JM, Rogers RJ, Tan WH. Anal Chem. 2005;77:4713. doi: 10.1021/ac050881y. [DOI] [PubMed] [Google Scholar]
- 38.Actis P, Maalouf MM, Kim HJ, Lohith A, Vilozny B, Seger RA, Pourmand N. ACS Nano. 2014;8:546. doi: 10.1021/nn405097u. [DOI] [PMC free article] [PubMed] [Google Scholar]