Figure 5.
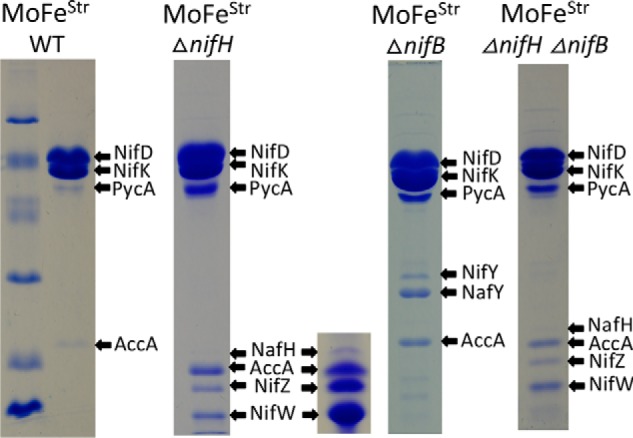
SDS-PAGE of Strep-tagged MoFe protein samples prepared from various genetic backgrounds using Streptactin affinity columns. Protein samples shown here and in other figures were separated using a 4% acrylamide stacking gel and 15% acrylamide running gel and then stained with Coomassie Brilliant Blue. Protein standards in the left panel include phosphorylase B (97.4 kDa), BSA (66.2 kDa), ovalbumin (45.0 kDa), carbonic anhydrase (31.0 kDa), soybean trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa). The inset located adjacent to the sample showing proteins that co-purify with MoFe protein produced in a ΔnifH background represents an overloaded sample to clearly show co-purification of NafH. Arrows indicate MoFe protein subunits and co-purifying nitrogen fixation-related proteins and also indicate prominent biotin-binding proteins unrelated to nitrogen fixation, acetate carboxylase subunit (AccA) and pyruvate carboxylase subunit (PycA), that are independently captured by the affinity-purification method. Higher levels of PycA and AccA in certain samples reflect the relatively lower abundance of MoFe protein subunits in extracts of those samples. Very light bands recognized in some samples were identified as various MoFe protein subunit degradation products. All proteins were identified by MS as described under “Materials and methods.” Neither MoFe protein nor any other nitrogen fixation-related protein binds to the Streptactin column if there is no Strep-tag on the MoFe protein.
