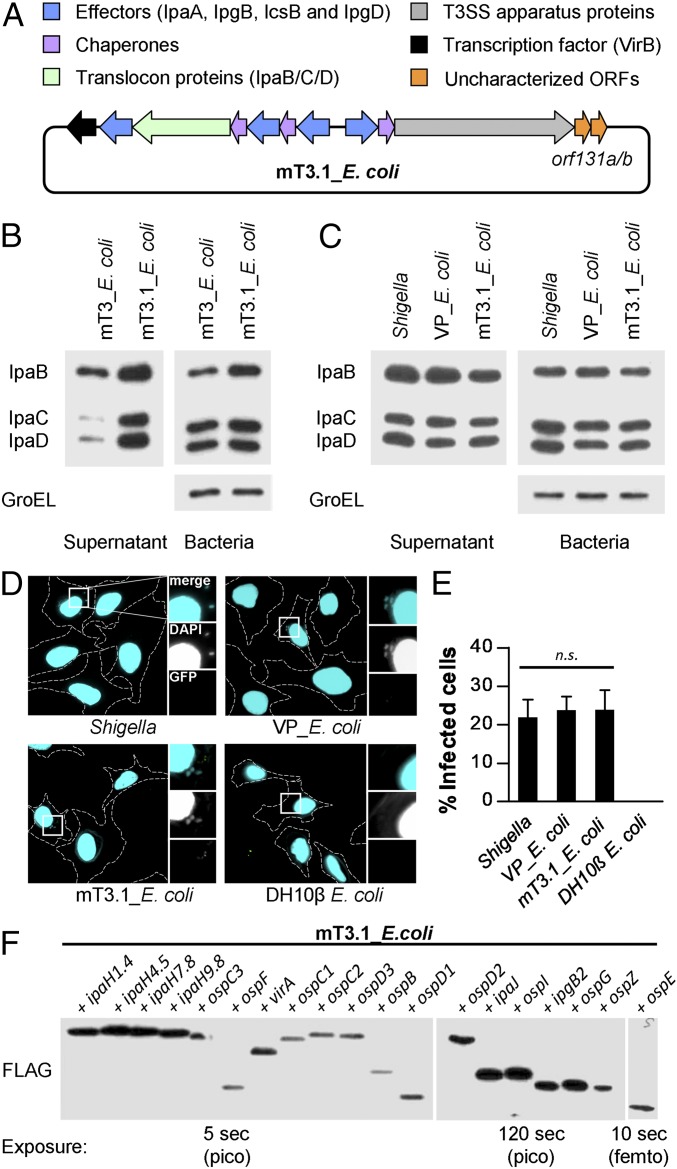
Fig. 1.
A synthetic bottom-up platform to study Shigella type III secreted effectors. (A) Schematic representations of the T3SS-encoding plasmid present in mT3.1_E. coli. (B and C) Secretion assays of designated strains. Each was grown under conditions that activate expression of its T3SS. A portion of the bacteria was removed and pelleted, and the remaining bacteria were resuspended in PBS plus Congo red, a condition that activates the machine. Proteins in the initial pellet and supernatant fractions collected at 30 min after the activation of secretion were separated by SDS/PAGE and transferred to nitrocellulose membranes. Membranes were immunoblotted with designated antibodies. GroEL, an unrelated cytosolic protein, served as a loading control. (D and E) HeLa cells were infected with each strain at an MOI of 100 for 30 min, followed by the addition of gentamicin to the media to kill extracellular bacteria. After 30 min, cells were labeled using an inside/outside microscopy assay to quantify intracellular (cyan) vs. extracellular (cyan/green) bacteria. (D) Representative images obtained with a 60× objective of labeled cells, with perimeters denoted by dashed lines. Enlarged images are of the boxed regions. (E) Quantification of infected cells. For each infection, 50 HeLa cells were counted. Data are expressed as the mean ± SD of three independent experiments. n.s., nonsignificant. (F) Secretion assays conducted as described in B and C. Membranes were immunoblotted with an antibody that recognizes the FLAG epitope. Multiple exposures were obtained to detect secretion of all effectors. femto, femtogram-level detection; pico, picogram-level detection. Blots in B, C, and F are representative of at least three experimental repeats.