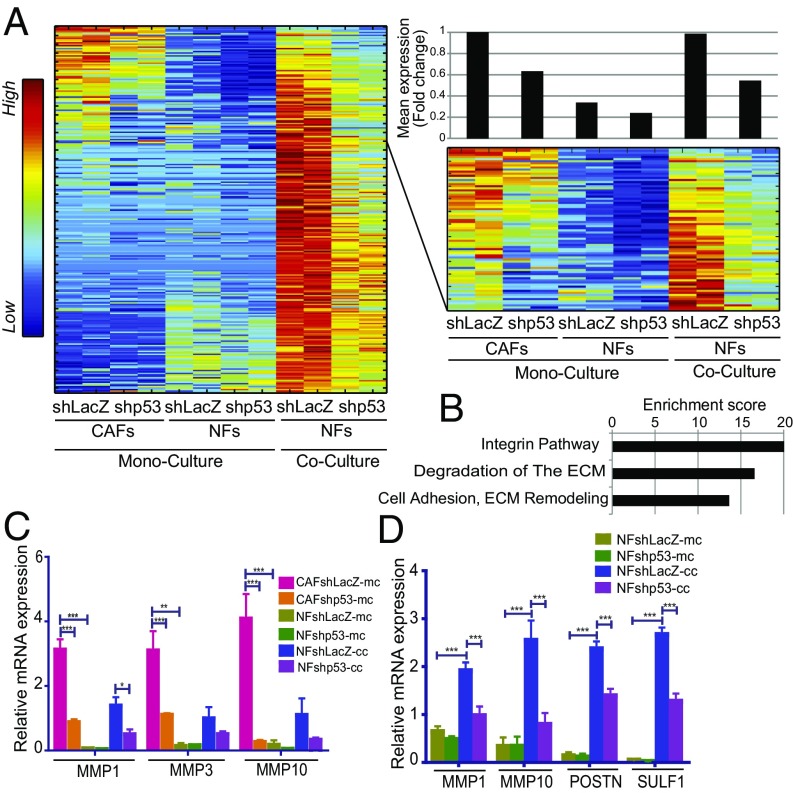
Fig. 6.
Tumor cells promote p53-dependent expression of CAF-associated genes in cocultured NFs. (A) SPIN-ordered expression matrix of genes up-regulated in a p53-dependent manner in immortalized NFs (patient 4731) cocultured with H460 cells, isolated by FACS sorting before RNA extraction. The upper portion of the heatmap is enlarged on the right side of the panel, to highlight the subset of genes also expressed highly in monocultured CAFs. Top Right shows mean fold-change relative to control (column 1) in each pair of replicate samples, calculated for the 77 genes shown in the enlarged panel. (B) Pathway enrichment analysis (GeneAnalytics) of the 77 genes in the right matrix in A. (C) Expression of MMP1, MMP3, and MMP10 mRNA in immortalized CAFs and NFs, grown as monoculture or cocultured with H460 cells and FACS-sorted before RNA extraction. The monoculture data are the same as in Fig. 3D and are included only for comparison. Values are means ± SEMs from four independent experiments. (D) Expression of MMP1, MMP10, POSTN, and SULF1 mRNA in immortalized NFs (patient 4731), grown as monoculture or cocultured with H1299 cells, and FACS-sorted before RNA extraction. Values are means ± SEMs from at least three independent experiments. *P ≤ 0.05, **P < 0.01, and ***P ≤ 0.001 using one-way ANOVA and Tukey post hoc test. SPIN, sorting points into neighborhood.