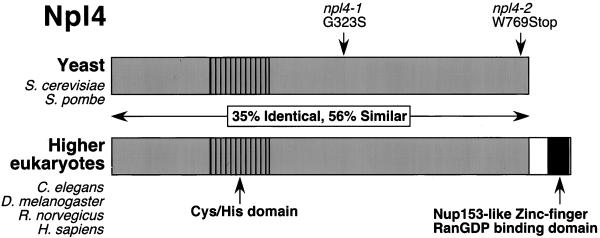
Figure 1.
The Npl4 protein is evolutionarily conserved. The sequence homology between S. cerevisiae Npl4p protein (ScNpl4p) (accession number CAA85131), S. pombe predicted protein SPBC1711.10c (SpNpl4) (accession number CAB88240), C. elegans proteins F59E12.4 (CeNpl4[1]) (accession number AAB54254) and F59E12.5 (CeNpl4[2]) (accession number AAB54255), D. melanogaster predicted protein CG4673 (DmNpl4) (accession number AAF56480), R. norvegicus (rat) Npl4 (Meyer et al., 2000), and predicted H. sapiens (human) protein FLJ20657 (HsNpl4) (accession number NP 060391) is depicted schemati-cally. The D. melanogaster Npl4 protein contains a large amino acid insertion omitted from this alignment. Ten absolutely conserved cysteine and histidine residues in the N terminus may constitute a novel Zn2+-finger domain. A C-terminal extension of higher eukaryotic Npl4 homologues encodes a predicted Nup153-like zinc-finger RanGDP-binding domain. The location of residues affected by mutations in the S. cerevisiae npl4-1 and npl4-2 mutants are indicated. The npl4-1 allele substitutes serine for the conserved glycine at position 323, and the npl4-2 mutation results in the production of a truncated protein lacking the C-terminal 12 amino acids.