Abstract
The FAD8 gene catalyzes the conversion of diene fatty acids to triene fatty acids and is a key enzyme that determines the synthesis of alpha-linolenic acid. In this study, the full-length cDNAs of FAD8-1, FAD8-2, and FAD8-3 are cloned from Paeonia ostii T. Hong & J. X. Zhang and named as PoFAD8-1, PoFAD8-2, and PoFAD8-3. Their open reading frame is 1203 bp, 1152 bp, and 1353 bp which encoded 400, 371, and 450 amino acids. The molecular weights of the amino acids are 46 kDa, 43 kDa, and 51 kDa while the isoelectric points are 7.34, 8.74, and 9.23, respectively. Bioinformatics analysis shows that all three genes are hydrophobic-hydrophobic, PoFAD8-1 has three transmembrane domains, and PoFAD8-2 and PoFAD8-3 have two transmembrane domains. Multiple series alignment and phylogenetic analysis revealed that PoFAD8-1 and PoFAD8-2 are closely related while PoFAD8-3 is more closely related to Paeonia delavayi. Subcellular localization results showed that PoFAD8-1 was located on the ER membrane and PoFAD8-2 and PoFAD8-3 were located on the chloroplast membrane. The relative expression level of PoFAD8-1 in seeds is very high. PoFAD8-2 expressed more in the ovary than the other two genes. PoFAD8-3 was highly expressed in roots, stems, leaves, petals, and ovaries.
Keywords: Paeonia ostii, FAD8, bioinformatics, quantitative real-time PCR, subcellular localization
1. Introduction
Fatty acid desaturase (FAD) is a key enzyme in plant lipid metabolism, which can promote the conversion of fatty acids to unsaturated fatty acids. At present, many genes involved in the process of fatty acid desaturase metabolism have been isolated from many plants. In model plant Arabidopsis thaliana, they are FAD2, FAD3, FAD6, FAD7, and FAD8 [1]. Since it is responsible for the catalytic material, it will be divided into two major categories of ω-3 and ω-6, which entails catalytic unsaturated fatty acid synthesis of fatty acid dehydrogenase [2,3]. The present studies show that FAD7 and FAD8 are all expressed in plastids, which mainly affect the composition of the thylakoid membrane lipids. So far, there are many studies on the function of FAD3 and FAD7 genes, but little research on the FAD8 gene [4,5,6]. The FAD8 gene was first found in Arabidopsis thaliana in 1994. It is located in the chloroplast of plant cells and uses ferredoxin as an electron donor to introduce a double bond at the omega-3 fatty acid position, which catalyzed the conversion of diene fatty acids (linoleic acid) to triene fatty acids (linolenic acid) [7]. FAD8 is also a key enzyme that determines the synthesis of alpha-linolenic acid (ALA) [8]. In Arabidopsis thaliana, FAD8 gene has been found to be constantly expressed at room temperature and its transcriptional regulation is easily induced by low temperatures [9]. Moreover, the physiological process of converting linoleic acid into linolenic acid can increase the degree of unsaturation of cell membrane lipids and it is one of the main ways for plants to adapt to low-temperature environments [10]. In addition to its association with low temperatures, FAD8 is also involved in the defense response mediated by jasmonic acid [11], ABA (abscisic acid), and SA (salicylic acid) [12]. FAD8 in tobacco also exhibits the anti-drought stress function [13] and corn FAD8 shows salt stress resistance [14].
Paeonia ostii T. Hong & J. X. Zhang, also known as Tongling peony, is mainly produced in Tongling County in the Anhui Province of China. Paeonia ostii T. Hong & J. X. Zhang belongs to Paeoniaceae and is the main species of peony oil. Peony seeds are wrapped in a star-shaped pod, which contains large amounts of unsaturated fatty acids (UFAs > 90%) and a high proportion of omega-3 fatty acids. These substances are associated with the prevention of various diseases such as cancer, cardiovascular disease, inflammation, and autoimmune diseases [15,16]. Studies have shown that FAD2 and FAD8 are highly expressed in peony seeds, which indicates that FAD2 and FAD8 may play a crucial role in forming polyunsaturated fatty acids. Among them, the relative expression level of FAD8 is much higher than FAD2 [17]. Li’s research shows that the high content of ALA in peony seeds is due to the activity and abundance of the FAD8 enzyme [17]. Therefore, as a new type of oilseed crop, it is necessary to study the expression pattern of the FAD8 gene in peony plants. At present, there are few reports on the expression pattern and function of the PoFAD8 gene in Paeonia ostii. We obtained three PoFAD8 genes by cloning from Paeonia ostii T. Hong & J. X. Zhang to explore the role of PoFAD8-1, PoFAD8-2, and PoFAD8-3.
2. Materials and Methods
2.1. Plant Material and Treatments
The experiment was conducted with the white peony cultivar Paeonia ostii T. Hong & J. X. Zhang planted in the Peony Germplasm Resources Park (32°23′ N, 119°24′ E) of Yangzhou University in Yangzhou, Jiangsu Province. The roots, stems, leaves, petals, ovaries, and seeds of 55 days, 70 days, 85 days, and 100 days after flowering were collected. At least three plant samples are repeated three times for each experiment. All samples were collected and placed in liquid nitrogen immediately for rapid freezing and then placed in a −80 °C cryogenic refrigerator to save them.
2.2. Extraction of Seed Oil and Measurement by GC-MS
The dried seed samples of different stages were peeled and crushed into powdered form. The oil content of developing seeds was extracted as described in our previous study [18]. Oil was extracted through an ultrasonic assisted extraction method. 30.0 g of seed powder was weighed and transferred into a 500 mL round bottom flask. Then n-hexane was mixed in with the material at a 1:13 ratio (g/mL). Afterwards, the flask was connected to a condenser tube and placed in the ultrasonic cleaning machine to perform an oil extraction at 70 °C and 200 W of power at a fixed frequency of 80 kHz. After extracting for 1.5 h, the mixture was centrifuged at 4000 r/min for 10 min. The supernatant was separated and dried by rotary evaporation at 50 °C for oil separation and n-hexane recovery.
The fatty acids (FAs) of Paeonia ostii seeds were extracted and were methylated according to procedures in the previous study [19]. FA content was analyzed through a gas chromatograph-mass (GC-MS) spectrometer (GC7890A/MS5975C, Agilent Technologies, Santa Clara, CA, USA) with HP-88 column (100 m × 0.25 mm i.d, 0.20-μm film thickness; Agilent). An internal standard curve was used to calculate unsaturated fatty acid content. The methyl heptadecanoate was used as the internal standard and the fatty acid methyl esters (FAMEs) were measured in each sample. The FAMEs of samples were recorded as milligrams per gram of dry weight (DW). All samples were analyzed in triplicate.
2.3. Cloning of Full-Length PoFAD8-1, PoFAD8-2, and PoFAD8-3 cDNA
The instructions of the TaKaRa MiniBEST Plant RNA Extraction Kit were used to extract the total RNA of the seeds and leaves. After RNA extraction, 1 μL was taken for agarose gel electrophoresis to measure its quality and concentration while the rest was stored at −80 °C in a refrigerator.
Using the designed primers to amplify the cDNAs of PoFAD8-1, PoFAD8-2, and PoFAD8-3, the amplified PCR products were detected by using 1% agarose gel electrophoresis. The target band was recovered by TaKaRa’s MiniBEST Agarose Gel cDNA Extraction Kit and then TA cloned using the pEASYTM-T5 Zero Cloning Kit. Then, the cloning vector was transformed into E. coli DH5α competent cells. Then after antibiotic (ampicillin) screening, the positive clones were finally sent to Shanghai Bioengineering Co., Ltd. (Shanghai, China) for sequencing.
2.4. Bioinformatic Analysis
The full-length open reading frame of cDNA and the encoded amino acids were predicted using Bioedit software. Biological software DNAMAN 5.0 was used to analyze translational sequences. The homology alignment of the NCBI Blast was carried out using the on-line tool NCBI Blast and the phylogenetic tree was constructed by using the NJ (Neighbor-joining) adjoining method of MEGA5.0 with the current major oil plants. The softwares Prot Param (http://web.expasy.org/protparam/), Hydrophobicity ProtScale (http://web.expasy.org/cgi-bin/protscale/protscale.pl), TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/), SMART (http://smart.embl-heidelberg.de/smart/), and SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) were used to analyze the physicochemical properties, hydrophilicity, and hydrophobicity as well as the transmembrane domains and secondary structure of the gene encoding proteins, respectively.
2.5. Protein Subcellular Localization
To further confirm the plastidial localization of the PoFAD8-1, PoFAD8-2, and PoFAD8-3 protein, full-length ORF PCR production of target genes that contain the BsaI/Eco31I restriction cut sites were cut by the endonuclease (Takara, Tokyo, Japan). Purified fragments were then ligated into the vector pBWA(V)HS-ccdb-GLosgfp. The pBWA(V)HS-ccdb-GLosgfp vector contains the Cauliflower mosaic virus (CaMV) 35S promoters and the mGFP reporter gene was fused with PoFAD8-1, PoFAD8-2, and PoFAD8-3. The p35S::PoFAD8-1-GFP, p35S::PoFAD8-2-GFP, and p35S::PoFAD8-3-GFPconstructs along with the empty pBWA(V)HS-GFP, vector pBWA(V)HS-ER-mKate vector (endoplasmic reticulum positioning signal), and pBWA(V)HS-Chloroplast-mKate vector (chloroplast positioning signal) were transformed into O. sativa protoplasts using a modified procedure described previously [20].
The rice seedlings were dark-cultured at 25 °C for 1 to 2 weeks. Fresh rice seedlings were cut into sections and added with the appropriate amount of enzyme solution (1.5% Cellulase R10, 0.75% Macerozyme R10, 0.6 M mannitol, 10 mM MES pH 5.7, 10 mM CaCl2, and 0.1% BSA). Gently shake (20–30 rpm) for 3 h at 15–20 °C. The collected protoplasts were transformed under PEG mixture [40% (w/v) PEG 4000, 2 M mannitol, and 0.1 M CaCl2] with 10 µg target plasmid. After incubate at room temperature for 16–24 h, the expression of plasmid was observed under a laser scanning confocal microscope.
2.6. Gene Expression Analysis
Using Bio-Rad’s CFX96 ReakTime System (Bio-Rad, Hercules, CA, USA), the gene sequences of PoFAD8-1, PoFAD8-2, and PoFAD8-3 were analyzed by qRT-PCR (real-time quantitative PCR) in the roots, stems, leaves, petals, ovary, and seed (60 d) of all parts of the seedlings and seeds of the 55 d, 70 d, 85 d, and 100 d expression differences. Real-time quantitative PCR was performed using the PrimeScript RT reagent Kit with gDNA Eraser kit (TaKaRa, Tokyo, Japan). The internal reference for the Paeonia ostii’s ubiquitin gene with specific expression primers shown in Table 1, have amplification conditions as follows: denaturation at 94 °C for 3 min, denaturation at 94 °C for 30 s, annealing at 54 °C for 30 s, extension at 72 °C for 1 min, 35 cycles, and extension at 72 °C for 10 min. The reaction Ct values were collected using Bio-Rad CFX Manager V1.6.541.1028 software. Repeat three times for each sample. The final result was calculated using the 2−ΔΔCt method [21].
Table 1.
Primers and sequences of PoFAD8-1, PoFAD8-2, and PoFAD8-3 gene cloning.
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Ubiquitin | GACCTATACCAAGCCGAAG | CGTTCCAGCACCACAATC |
| qFAD8-1 | ACCTTAGAGGAGGGCTTACGACAAT | CGTAGTGGTCTTGCTTGATGCTCCT |
| qFAD8-2 | TAGTGGCATGAAGATGAATCAAGAT | CGCTTGCTATTAGTCCCAGAACCAC |
| qFAD8-3 | ATGATAGCCCCTCAAAGAATAGAAT | CGCAACATCCCTCACAACATAGC |
2.7. In Vitro Expression of FAD8-1 through Cell-Free Expression System
The expression plasmid of PoFAD8-1 was constructed. The full-length ORF of PoFAD8-1 was amplified and connected onto pET21a vector, which was promoted by the T7 promoter and labeled with a 10 xhis tag. Additionally, 30 μL mixture contained E. coli extracts, feed buffers, amino acids, and T7 RNA polymerase from the Expressway™ Mini Cell-Free Expression System (Thermo Fisher, Waltham, MA, USA) and 20 μL plasmid pET21a-PoFAD8-1 (concentration at 150 μg/mL) was added into the dialysis box. Then the cassette was immersed in the feed buffer at 30 °C overnight. The reaction solution was taken and centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatant was discarded and the precipitates were diluted and detected by SDS-PAGE.
3. Results
3.1. Cloning and Nucleotide Sequence Analysis of PoFAD8-1, PoFAD8-2, and PoFAD8-3
The cDNA fragments of PoFAD8-1, PoFAD8-2, and PoFAD8-3 were obtained by PCR amplification from Paeonia ostii T. Hong & J. X. Zhang. After sequencing the PCR products, their sequences were contrasted with FAD genes of Arabidopsis thaliana and blasted at Tair (http://www.arabidopsis.org/). Afterward, they were identified as PoFAD8-1, PoFAD8-2, and PoFAD8-3. Their sequences were uploaded onto NCBI and their accession numbers were MH049427, MH049428, and MH049429, respectively. Sequence analysis showed that the ORF of the PoFAD8-1 gene was 1203 bp in length and encoded 400 amino acids with a molecular weight of 46 kDa and an isoelectric point of 7.34. The ORF of the PoFAD8-2 gene was 1152 bp in length and encoded 371 amino acids. The molecular weight of the gene was 43 kDa and the isoelectric point was 8.74. The ORF of PoFAD8-3 gene is 1353 bp in length and encodes a protein of 450 amino acids with a molecular weight of 51 kDa and an isoelectric point of 9.23. Genomic DNA was amplified by PCR using primers PoFAD8-1, PoFAD8-2, and PoFAD8-3.
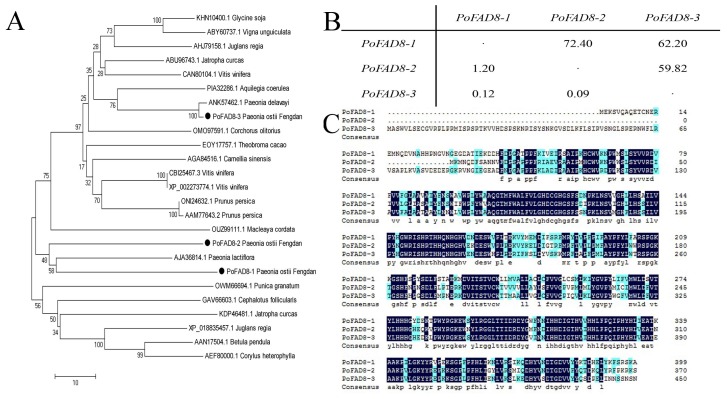
To compare the differences of the sequence of PoFAD8-1, PoFAD8-2, and PoFAD8-3 (Supplementary Figure S1), DNAMAN software was used to compare the amino acid composition and the number of PoFAD8-1, PoFAD8-2, and PoFAD8-3 genes with the protein sequence, which is shown in Table 2 and Figure 1C. MEGA software was used to compare the amino acid sequences of PoFAD8-1, PoFAD8-2, and PoFAD8-3 with the FAD8 gene of 17 species. The Neighbor-Joining system is constructed to set the number of bootstrap to be 1000 and the results are shown in Figure 1A. Cluster analysis showed that PoFAD8-1 was closely related to PoFAD8-2. PoFAD8-3 is more closely related to Paeonia delavayi. PoFAD8-1 and PoFAD8-2 shared a close evolutionary relationship with Paeonia lactiflora and PoFAD8-3 shared close evolutionary origins with Aquilegia coerulea. The PoFAD8-1 and PoFAD8-2 genes are clustered in different regions with PoFAD8-3, which indicates that the PoFAD8-1 and PoFAD8-2 genes are more distantly related to PoFAD8-3. DNAMAN was used to do the pairwise alignment of three genes. The pairwise comparison of PoFAD8-1, PoFAD8-2, and PoFAD8-3 with MEGA for nucleic acid molecular evolution analysis show that PoFAD8-1 and PoFAD8-2 genes have high identity and their identity rate with PoFAD8-3 is low. Ka/Ks values of PoFAD8-1 and PoFAD8-2 are close to 1, which indicates that these two genes have not received or seldom been under the pressure of natural selection and they are moderately evolved. The Ka/Ks values of PoFAD8-1 and PoFAD8-3 as well as PoFAD8-2 and PoFAD8-3 are far less than 1. This can be explained by the gene being subject to purification. The results are shown in Figure 1B.
Table 2.
Amino acid composition and number of products encoded by cDNAs of PoFAD8-1, PoFAD8-2, and PoFAD8-3 gene.
| Amino Acids | PoFAD8-1 | PoFAD8-2 | PoFAD8-3 | Amino Acids | PoFAD8-1 | PoFAD8-2 | PoFAD8-3 |
|---|---|---|---|---|---|---|---|
| Ala (A) | 22 | 14 | 24 | Leu (L) | 30 | 31 | 40 |
| Arg (R) | 14 | 16 | 24 | Lys (K) | 22 | 17 | 24 |
| Asn (N) | 15 | 14 | 20 | Met (M) | 11 | 12 | 9 |
| Asp (D) | 22 | 19 | 22 | Phe (F) | 23 | 22 | 20 |
| Cys (C) | 7 | 4 | 5 | Pro (P) | 23 | 28 | 31 |
| Gln (Q) | 7 | 8 | 5 | Ser (S) | 25 | 21 | 38 |
| Glu (E) | 17 | 10 | 18 | Thr (T) | 18 | 16 | 17 |
| Gly (G) | 25 | 23 | 30 | Trp (W) | 13 | 15 | 16 |
| His (H) | 28 | 24 | 28 | Tyr (Y) | 23 | 21 | 20 |
| Ile (I) | 20 | 22 | 20 | Val (V) | 35 | 34 | 39 |
Figure 1.
Multi-series alignment and phylogenetic analysis. (A) Neighbor-Joining System evolutionary tree of PoFAD8-1, PoFAD8-2, and PoFAD8-3 genes with another 17 species. The GenBank accession numbers are: Glycine soja (GsFAD8, KHN10400.1), Vigna unguiculata (VuFAD8, ABY60737.1), Juglans regia (JrFAD8, AHJ79158.1), Jatropha curcas (JcFAD8, ABU96743.1) Vitis vinifera (VvFAD8, CAN80104.1), Aquilegia coerulea (AcFAD8, PIA32286.1), Paeonia delavayi (PdFAD8, ANK57462.1), Corchorus olitorius (CoFAD8, OMO97591.1), Theobroma cacao (TcFAD8, EOY17757.1), Camellia sinensis (CsFAD8, AGA84516.1), Vitis vinifera (VvFAD8, CBI25467.3), Vitis vinifera (VvFAD8, XP_002273774.1), Prunus persica (PpFAD8, ONI24632.1), Prunus persica (PpFAD8, AAM77643.2), Macleaya cordata (McFAD8, OUZ99111.1), Paeonia lactiflora (PlFAD8, AJA36814.1), Punica granatum (PgFAD8, OWM66694.1), Cephalotus follicularis (CfFAD8, GAV66603.1), Jatropha curcas (JcFAD8, KDP46481.1), Juglans regia (JrFAD8, XP_018835457.1), Betula pendula (BpFAD8, AAN17504.1), and Corylus heterophylla (ChFAD8, AEF80000.1). The horizontal scale shows the difference number per 100 residues from the Clustal W alignment. (B) Comparisons of PoFAD8 paralogues. Ka/Ks and amino acid sequence identity (%) values for pairwise comparisons of PoFAD8 paralogues are shown. Ka/Ks values are shown below the diagonal while amino acid sequence identity (%) values are shown above the diagonal. (C) Protein sequence alignment of PoFAD8-1, PoFAD8-2, and PoFAD8-3 genes. The blue black and light blue boxes show the same and similar amino acids, respectively.
3.2. Bioinformatic Analysis of the PoFAD8-1, PoFAD8-2, and PoFAD8-3 Protein
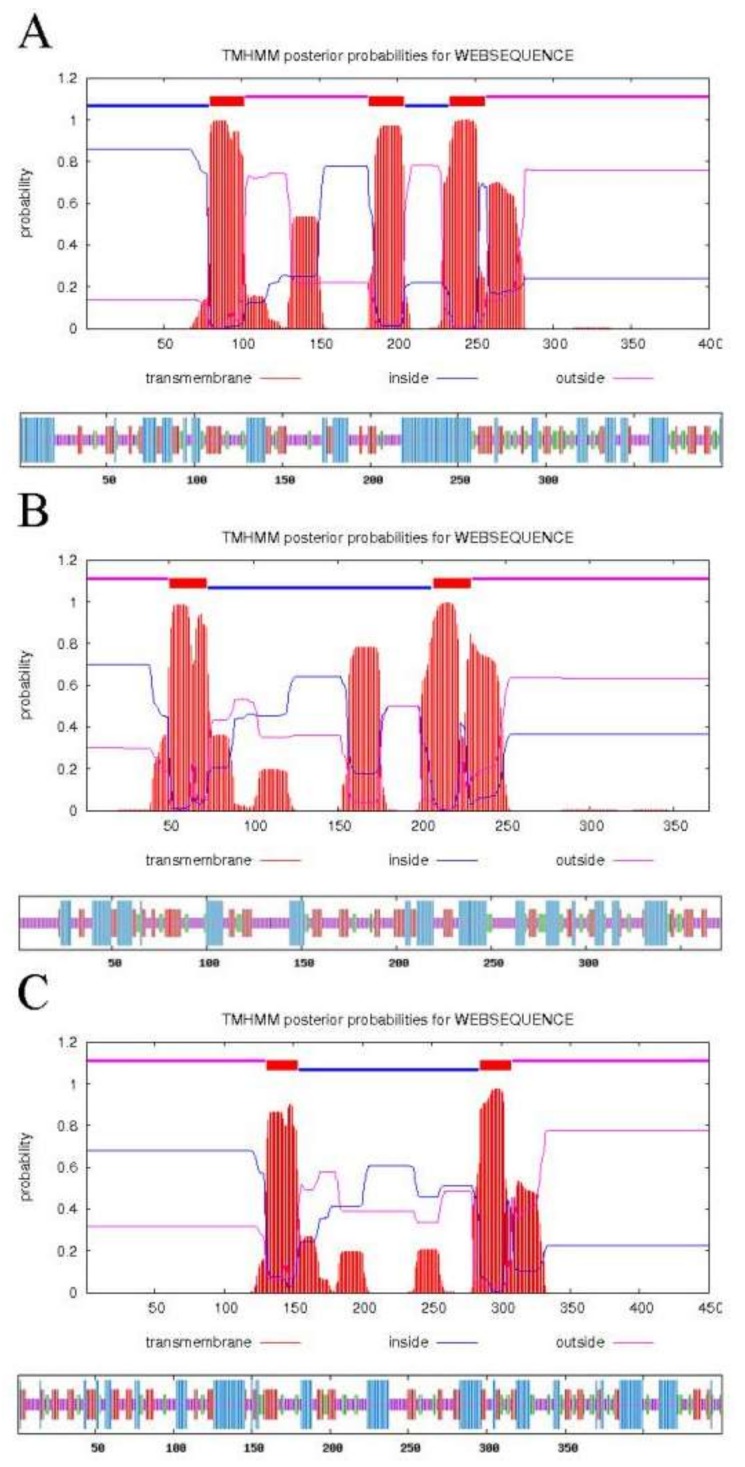
The analysis results are shown in Figure 2 below. It can be seen from Figure 2A that PoFAD8-1 has three predicted transmembrane domains of 80 to 102, 182 to 204, and 234 to 256 amino acids. The protein encoded by PoFAD8-1 has a minimum hydrophobicity of −2.867 and a maximum of 3.267, which indicates that the protein is a hydrophobic protein. In the secondary structure prediction of PoFAD8-1, the total alpha helix has 140 amino acids, which accounts for 35%. The total extended strand is 80 amino acids, which accounts for 20%. The total random coil is 131 amino acids, which accounts for 32.75%. The total beta turn is 49 amino acids, which accounts for 12.25%. The alpha helix, extended strand, random coil, and beta turn run through the entire amino acid chain. In Figure 2B, PoFDA8-2 has two transmembrane domains of 50 to 72 and 207 to 229 amino acids. In addition, the protein encoded by the PoFAD8-2 had a minimum hydrophobicity of −2.867 and a maximum of 3.433, which indicates that the protein is a hydrophobic protein. The secondary structure of PoFAD8-2 predicted that the total alpha helix has 105 amino acids, which accounts for 28.3%. The total extended strand has 79 amino acids, which accounts for 21.29%. The total random coil has 150 amino acids, which accounts for 40.43%. The total beta turn has 37 amino acids, which accounts for 9.97%. The alpha helix, extended strand, random coil, and beta turn run through the entire amino acid chain. In Figure 2C, PoFAD8-3 has two transmembrane domains of amino acids 131 to 153 and 285 to 307, respectively. The protein encoded by the PoFAD8-3 gene has a minimum hydrophobicity of −2.867 and a maximum of 3.211, which indicates that the protein is a hydrophobic protein. In the secondary structure prediction of PoFAD8-3, the total alpha helix has 126 amino acids, which accounts for 28%. The total extended strand has 101 amino acids, which accounting for 22.44%, and the total random coil has 163 amino acids accounting for 36.22%. Additionally, the total beta turn has 60 amino acids, which accounts for 13.33%. The alpha helix, extended strand, random coil, and beta turn run through the entire amino acid chain.
Figure 2.
Transmembrane domain prediction and secondary structure prediction of PoFAD8-1, PoFAD8-2, and PoFAD8-3. (A) Transmembrane domain prediction and secondary structure prediction of PoFAD8-1. (B) Transmembrane domain prediction and secondary structure prediction of PoFAD8-2. (C) Transmembrane domain prediction and secondary structure prediction of PoFAD8-3.
3.3. Subcellular Localization of the PoFAD8-1, PoFAD8-2, and PoFAD8-3 Protein
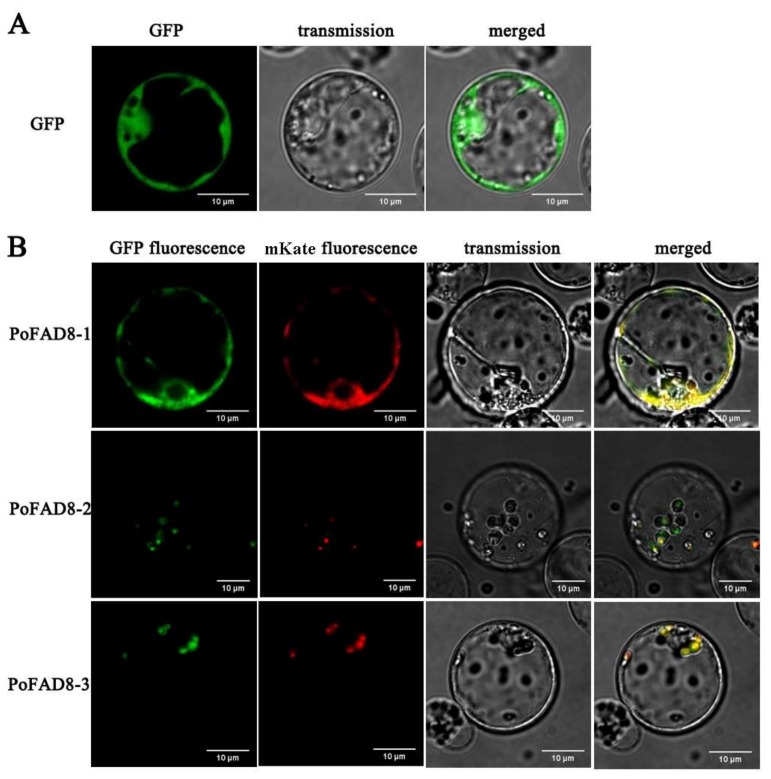
To determine the subcellular localization of the PoFAD8-1, PoFAD8-2, and PoFAD8-3 protein, the fusion protein expression vector pBWA(V)HS-FAD8-1-GFP, pBWA(V)HS-FAD8-2-GFP, and pBWA(V)HS-FAD8-3-GFP were transformed into the protoplast of rice by the PEG method using an empty vector pBWA(V)HS-GFP as a control. The pBWA(V)HS-ER-mKate vector (endoplasmic reticulum positioning signal) and vector pBWA(V)HS-Chloroplast-mKate vector (chloroplast positioning signal) were used as a subcellular co-localization vector.
Using the GFP empty vector as a positive control, transgenic rice protoplasts were stimulated by 488 nm to emit green fluorescence. As shown in Figure 3A, the green fluorescence sites were widely distributed in the whole plasma membrane system and nucleus but not in the vacuole (central dark part) and chloroplast. Figure 3B showed that PoFAD8-1 only appeared on the endoplasmic reticulum membrane and was consistent with the localization of mKate. This result indicated that PoFAD8-1 was located on the endoplasmic reticulum membrane. The fluorescence intensity of the PoFAD8-1-GFP vector (B) was less than the empty vector control. However, the green fluorescence of the 35S::FAD8-2-GFP and 35S::FAD8-3-GFP chimeras were located at the chloroplast membrane, which was co-localized with chloroplast-mKate in the protoplasts. These results indicated that PoFAD8-1 was an ER membrane protein, which was consistent with subcellular localization prediction results and was different from the other two family members PoFAD8-2 and PoFAD8-3.
Figure 3.
Subcellular localization of PoFAD8-1, PoFAD8-2, and PoFAD8-3 in rice protoplasts.
3.4. Expression Pattern of the PoFAD8-1, PoFAD8-2, and PoFAD8-3 Genes
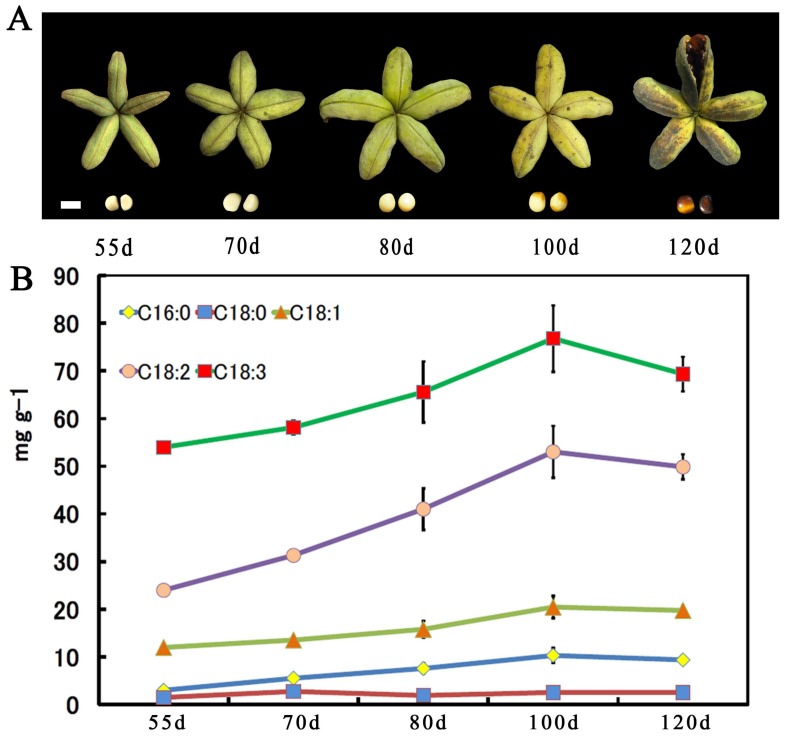
The seed development process was observed from 55 days after pollination until 120 days maturation. The pods of Paeonia ostii were hand-collected at approximately fifteen-day intervals from the 55 days after pollination DAP until full maturity, which covers a total range of 65 days including 55 days, 70 days, 85 days, 100 days, and 120 days (a total of five time points). Figure 4A shows that the size and color varied dramatically at different stages of seed development. The fatty acid contents of seeds from five developmental stages of Paeonia ostii were characterized by GC–MS and the FA content is depicted in Figure 4B. The results showed that five dominant components were found. These are called α-linolenic acid (C18:3Δ9c, 12c, 15c), linoleic acid (C18:2Δ9c, 12c), oleic acid (C18:1Δ9c), stearic acid (C18:0), and palmitic acid (C16:0). The combined content of these five FAs was more than 99.3% of total FAs at 100 days and it was always predominant across the seed developmental stages. The high proportion of n-3 FAs is quite rare in oil crops. Other minor FAs (<1.0%) were also detected including myristic acid (C14:0), palmitoleic acid (C16:1Δ9c), cis-11-octadecenoic acid (C18:1Δ11c), eicosanoic acid (C20:0), and cis-11-eicosenoic acid (C20:1Δ11c).
Figure 4.
Observation and measurement of lipids across the developmental period of Paeonia ostii seeds. (A) The developmental progress of Paeonia ostii seeds. The pods were harvested at 55 days after pollination (DAP, immature stage) and every approximately 15 days thereafter until 120 DAP (pods containing mature seeds), bar = 1 cm. (B) The total fatty acid content at five time points during Paeonia ostii seed development (mean ± SD, n = 3).
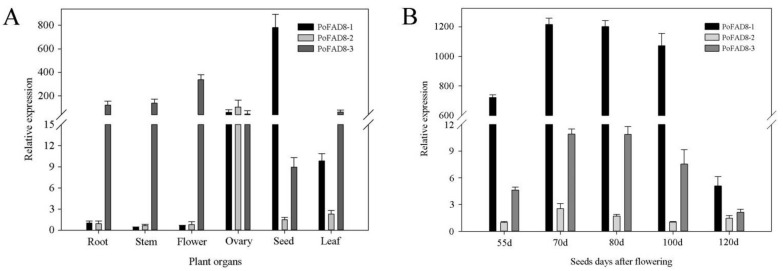
We measured the expression of PoFAD8-1, PoFAD8-2, and PoFAD8-3 in the roots, stems, leaves, petals, ovaries, and seeds of the genus Paeonia ostii T. Hong & J. X. Zhang by qRT-PCR. From Figure 5A, we can see that the expression of PoFAD8-1 in the ovary and seed is extremely high. The relative expression level of PoFAD8-1 in seeds was 781, which was 531 and 87 times of that of PoFAD8-2 and PoFAD8-3, respectively. PoFAD8-1 also has the highest expression among three genes in the measured six organs. PoFAD8-2 was expressed higher in the ovary than in the other two genes and its relative expression was 105. The expression of PoFAD8-3 in roots, stems, leaves, petals, and ovaries was high. Among them, the expression of PoFAD8-3 was far ahead of the other two genes in roots, stems, leaves, and petals and the highest relative expression level in flowers was 338, which were 553 and 427 fold of PoFAD8-1 and PoFAD8-2, respectively. Its expression in the seeds is higher than PoFAD8-2.
Figure 5.
PoFAD8-1, PoFAD8-2, and PoFAD8-3 genes expression were measured during seed development and different organs in the Paeonia ostii T. Hong & J. X. Zhang. (A) Tissue specific expression of PoFAD8-1, PoFAD8-2, and PoFAD8-3. (B) PoFAD8-1, PoFAD8-2, and PoFAD8-3 expression during Paeonia ostii seed development. The error bar represents the standard deviation of three replicates. The tree peony UBQ was used as an internal control.
From Figure 5B, we can see that during the five stages of seed development, PoFAD8-1 is always expressed the most and is followed by PoFAD8-3 and PoFAD8-2. The expression of PoFAD8-1 in the first four periods has been much higher than that of PoFAD8-2 and PoFAD8-3. The fifth period was slightly higher than PoFAD8-2 and PoFAD8-3. The trends of the five developmental stages of the three genes were similar. The 55 days, 70 days, and 80 days during these three periods, the expression of three genes increased. Interestingly, the relative expression of PoFAD8-1 decreased slightly at 100 days. In the 120 days, however, it dropped sharply from 1073 to 5. It is speculated that PoFAD8-1 may play a major function in ovaries and seeds, which specifically regulate unsaturated fatty acid synthesis. According to the GC-MS measurement of FA content, the highest content of ALA in 85 days may be caused by a high expression of PoFAD8-1. The relative expression of PoFAD8-1 dramatically decreased after 100 days. This is consistent with the decrease of Paeonia ostii unsaturated fatty acids after 100 days.
3.5. In Vitro Expression of PoFAD8-1 Gene
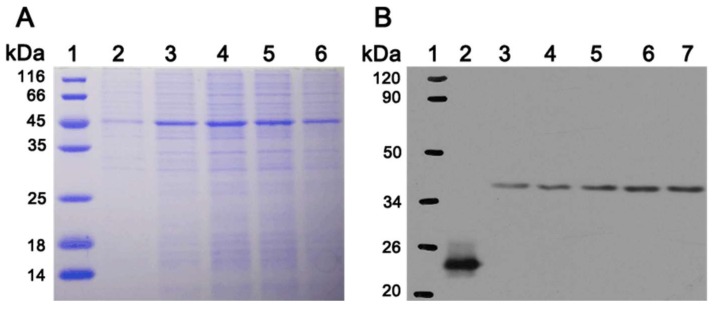
Because the subcellular localization of PoFAD8-1 is different from PoFAD8-2 and PoFAD8-3 and its expression level is always t its highest state, it indicated that it may be closely related to the high ALA content of peony seeds. Therefore, the PoFAD8-1 protein was heterologous expressed for further exploring the characteristics. In the earlier period, we used the Pichia pastoris expression system, but since PoFAD8-1 is a multi-spanning membrane protein, the in vitro secretion is very difficult. We did not obtain destination band in SDS-PAGE. After improvement, we constructed the in vitro expression of PoFAD8-1 driven by using the T7 promoter. We used cell-free expression technology and successfully expressed and purified the PoFAD8-1 protein. Two-fold dilutions of the cell-free expression system were loaded as 10 μL, 12 μL, 14 μL, 16 μL, and 18 μL (see Figure 6A). After SDS-PAGE and detection, the band size was 45 kDa and was consistent with the prediction. Following a western blot experiment, the results showed that the positive control obtained the correct imprinting of 23 kD and the size of PoFAD8-1 band was the same as the predicted 45 kD (see Figure 6B). After gel purification and mass spectrometry, the results showed that the amino acid sequence was identical to that of PoFAD8-1. It was demonstrated that the PoFAD8-1 in vitro expression experiment was successful.
Figure 6.
(A) The FAD8-1 protein was synthesized in vitro using the ExpresswayTM Mini Cell-Free Expression System. Products were loaded onto an SDS-PAGE gel and proteins were then visualized with Coomassie Blue staining. (B) Western Blot was also carried out and detected by his antibody. The exogenous positive control protein containing his tag at 23 kDa was used as a control.
4. Discussion
At present, there are relatively few studies on the function of FAD8 genes and they mainly focus on the related gene cloning and expression pattern analysis. Olga PY et al. [22] analyzed the ω-3 fatty acid desaturase gene family of Gossypium spp. from the genome level. The transcriptome sequencing revealed that FAD7/8-1 gene could be induced by low temperature. Similarly, a study of the CsFAD8 gene expression pattern in Camellia sinensis showed that the expression of this gene was induced by a low temperature and ABA stress. In addition, functional studies of Arabidopsis At-FAD8 proved that the gene can regulate the fatty acid content decreased with increasing temperature. However, Susan et al. [7] found that the FAD8 gene in Arabidopsis was induced by cold and expressed in large quantities below 20 °C, which rapidly increased the cold resistance of tissues [23]. Studies on the omega-3 fatty acid dehydrogenase gene in Glycine max (Linn.) Merr. showed that hypothermia did not affect the expression levels of GmFAD8-1 and GmFAD8-2 [24]. The study of the expression patterns of CmsFAD2 and CmsFAD8 genes in both cryopreserved and cold-sensitive Citrus medica L. var. sarcodactylis. Swingle showed that the expression level of the two genes correlated with the genotypes of the two plants [25,26]. In Arabidopsis, two types of omega-3 FADs (FAD3 located in the ER and FAD7/8 located in the chloroplast) catalyze the formation of TA [7,26,27]. Matilda et al. [23] found that, even though the two kinds of ω-3 fatty acid desaturase FAD7 and FAD8 in Arabidopsis have a high degree of structural homology, the response mechanisms of the two enzymes to the temperature are completely different. They transferred a series of constructed FAD7-FAD8 fusion genes into Arabidopsis FAD7-FAD8 double mutants. The effect of each gene on temperature response was examined by Northernm and Southern analysis and fatty acid composition measurements. The results showed that all the transgenic plants transfected into the construct with the FAD8 C-terminal coding region (44 amino acids) showed a significant decrease in TA levels at high temperature. This is similar to the plants obtained by transgenes with the FAD8 native structural gene. The expression profile of omega-3 FAD gene in Perilla frutescens was studied by transcriptome analysis. The results showed that FAD3 and FAD7/8 were the key genes for ALA synthesis in seeds and leaves, respectively [28]. The expression level of PfFAD7/8 in Perilla frutescens leaves was higher than that in seeds, which indicates that they play a preferential role in ALA accumulation in vegetative organs. After heat treatment of Perilla frutescens leaves for 48 hours, the transcripts of Shia ShFAD3-1/-2 and ShFAD7 slightly decreased and the expression of ShFAD8 slightly increased, which indicates that ShFAD8 might respond to heat [29].
In Arabidopsis thaliana, FAD7 and FAD8 are genes with similar functions. However, FAD8 is highly expressed under low temperature induction [7]. Its function is to catalyze the conversion of plant diene fatty acids (palmitate and linoleic acid) into TAs in the chloroplast. Post-transcriptional regulation of FAD8 exists and is closely related to the rapid increase of TAs at low temperature and the decrease of TAs at high temperature [23]. As can be seen from the molecular phylogenetic tree, the catalytic TAs-containing fatty acid desaturase gene is a large family of FAD7, FAD8, and plastid-expressed FAD3 [30]. In most plants, FAD7 and FAD8 have a closer genetic relationship. Both FAD7 and FAD8 have similar catalytic functions and different expression and regulation. In phylogenetic tree analysis, the two genes apparently become two branches, but the distances between different plants are larger. In general, the similarity between the two genes FAD7 and FAD8 is generally lower than that of FAD8 among different species.
Peony seed oil contains a lot of unsaturated fatty acids and gradually favored by consumers. The abundant α linolenic acid (C18:3) brings a great nutrition to human health. FADs, known as the fatty acid desaturase, control the key steps of oleic acid transfer to the linoleic acid and α linolenic acid. The regulation mechanism of synthesis of α linolenic acid is still unclear.
Generally FAD3, FAD7, and FAD8 are not a saturated fatty acid catalytic enzyme. Plants use them as key enzymes to produce C18:3. FAD3 is widespread distribution in endoplasm momentum while FAD7 and FAD8 distribution on the chloroplast membrane. In this study, the results show that, compared with FAD8-2 and FAD8-3, FAD8-1 changed the distribution of the position. However, the subcellular localization experiment proves its location is on the ER instead of the chloroplast membrane. The results suggest that during the evolution of peony, the functional differentiation of FAD8-1 happened. The subcellular level localization has also changed due to the area being widely distributed in the endoplasmic reticulum in cells. The cell-free protein expression system, also known as the in vitro translation system, simulates the life phenomena of biological cells and reproduces the translation and translation process of intracellular proteins. The cell-free protein synthesis system uses an external target mRNA or DNA as a template and artificially supplements the substrates required for protein synthesis and transcription and translation-related factors to achieve the in vitro synthesis of the target protein. PoFAD8-1 is used as a catalytic enzyme that is embedded in the membrane of the endoplasmic reticulum. After purification in vitro, how to make it form the correct spatial conformation and how to perform certain enzyme activities, the laboratory still needs further study.
5. Conclusions
In conclusion, we cloned the fatty acid desaturase genes PoFAD8-1, PoFAD8-2, and PoFAD8-3 of Paeonia ostii T. Hong & J. X. Zhang. A subcellular localization analysis confirmed that PoFAD8-1 is located on the ER membrane while PoFAD8-2 and PoFAD8-3 are located on the chloroplast membrane. The relative expression level of PoFAD8-1 in seeds was very high, which is also the highest expression level of the three genes in the tested six loci. PoFAD8-2 expresses more in the ovary than the other two genes. PoFAD8-3 is highly expressed in roots, stems, leaves, petals, and ovaries. In the five stages of seed development, the expression levels of PoFAD8-1 in the first four periods were much higher than those in PoFAD8-2 and PoFAD8-3 while the fifth period was slightly higher than PoFAD8-2 and PoFAD8-3. We speculate from these results that PoFAD8-1 has a very close relationship to high content of ALA in peony seed oil. Further, PoFAD8-1 in vitro expression experiments using the cell-free expression system were successfully performed and allowed other FAD8 members to be expressed in vitro. Since the endoplasmic reticulum is widely distributed in cells, it can provide a high catalytic efficiency for dehydrogenation of fatty acids. The change of PoFAD8-1 distribution in cells expanded its catalytic range, which made a contribution toward the high content of ALA in peony seeds. We believe that this report will provide a lot of information for further study of the peony FAD family and provide reference for the production of high ALA content of germplasm resources in the future.
Acknowledgments
This work was supported by funding from the National Natural Science Foundation of China (31600564), the Natural Science Fund of Jiangsu Province (BK20160460), the Three New Project for Agriculture of Jiangsu Province (Grant No. LYSX[2016]45), Compliance with Ethical Standards.
Supplementary Materials
The following are available online, Figure S1: Sequence alignment of PoFAD8-1, PoFAD8-2, and PoFAD8-3.
Author Contributions
Jun Tao and Jing Sun conceive and design experiments. Ming Chen conducts experiments. Mengyuan Zhu and Yu Jiang analyze data. Jiasong Meng and Daqiu Zhao write articles.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Cao F.L., Wang H.L., Yu W.W., Cheng H. Advances in fatty acid desaturase and genes of higher plants. J. Nanjing For. Univ. Natl. Sci. Ed. 2012;36:125–132. [Google Scholar]
- 2.Yazawa H., Iwahashi H., Kamisaka Y., Kimura K., Aki T., Ono K., Uemura H. Heterologous production of dihomo-gamma-linolenic acid in Saccharomyces cerevisiae. Appl. Environ. Microb. 2007;73:6965–6971. doi: 10.1128/AEM.01008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ailhaud G., Massiera F., Weill P., Legrand P., Alessandri J.M., Guesnet P. Temporal changes in dietary fats: Role of n − 6 polyunsaturated fatty acids in excessive adipose tissue development and relationship to obesity. Prog. Lipid Res. 2006;45:203–236. doi: 10.1016/j.plipres.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Liu X.Y., Teng Y.B., Li B., Meng Q.W. Enhancement of low-temperature tolerance in transgenic tomato plants overexpressing Lefad7, through regulation of trienoic fatty acids. Photosynthetica. 2013;51:238–244. doi: 10.1007/s11099-013-0014-5. [DOI] [Google Scholar]
- 5.Wang H.S., Yu C., Tang X.F., Zhu Z.J., Ma N.N., Meng Q.W. A tomato endoplasmic reticulum (ER)-type omega-3 fatty acid desaturase (LeFAD3) functions in early seedling tolerance to salinity stress. Plant Cell Rep. 2014;33:131–142. doi: 10.1007/s00299-013-1517-z. [DOI] [PubMed] [Google Scholar]
- 6.Tian E., Zeng F., Mackay K., Roslinsky V., Cheng B. Detection and molecular characterization of two fad3 genes controlling linolenic acid content and development of allele-specific markers in yellow mustard (sinapis alba) PLoS ONE. 2014;9:e97430. doi: 10.1371/journal.pone.0097430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson S., Arondel V., Iba K., Somerville C. Cloning of a temperature-regulated gene encoding a chloroplast omega-3 desaturase from Arabidopsis thaliana. Plant Physiol. 1994;106:1615–1621. doi: 10.1104/pp.106.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L.X., Tang G.Y., Chen G., Bi Y.P., Shan L. Polymorphism of AhFAD2 gene in peanut and its correlation with grain oleic acid/linoleic acid ratio. Acta Agron. Sin. 2011;37:415–423. doi: 10.3724/SP.J.1006.2011.00415. [DOI] [Google Scholar]
- 9.Somerville C. Direct tests of the role of membrane lipid composition in low-temperature-induced photoinhibition and chilling sensitivity in plants and cyanobacteria. Proc. Natl. Acad. Sci. USA. 1995;92:6215–6218. doi: 10.1073/pnas.92.14.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iba K. Abiotic Stress Tolerance in Plants. Springer Netherlands; Dordrecht, The Netherlands: 2006. Trienoic fatty acids and temperature tolerance of higher plants; pp. 61–68. [Google Scholar]
- 11.Browse J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry. 2009;70:1539–1546. doi: 10.1016/j.phytochem.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the arabidopsis ssi2 mutant. Plant Cell. 2003;15:2952–2965. doi: 10.1105/tpc.017301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M., Barg R., Yin M.G., Gueta-Dahan Y., Leikin-Frenkel A., Salts Y., Shabtai S., Ben-Hayyim G. Modulated fatty acid desaturation via overexpression of two distinct omega-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 2005;44:361–371. doi: 10.1111/j.1365-313X.2005.02536.x. [DOI] [PubMed] [Google Scholar]
- 14.Berberich T., Harada M., Sugawara K., Kodama H., Iba K., Kusano T. Two maize genes encoding omega-3 fatty acid desaturase and their differential expression to temperature. Plant Mol. Biol. 1998;36:297–306. doi: 10.1023/A:1005993408270. [DOI] [PubMed] [Google Scholar]
- 15.Simopoulos A.P. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. 2002;56:365–379. doi: 10.1016/S0753-3322(02)00253-6. [DOI] [PubMed] [Google Scholar]
- 16.Simopoulos A.P. Evolutionary aspects of diet, the omega-6/omega-3 ratio and genetic variation: Nutritional implications for chronic diseases. Biomed. Pharmacother. 2006;60:502–507. doi: 10.1016/j.biopha.2006.07.080. [DOI] [PubMed] [Google Scholar]
- 17.Li S.S., Wang L.S., Shu Q.Y., Wu J., Chen L.G., Shao S., Yin D.D. Fatty acid composition of developing tree peony (paeonia section moutan DC.) seeds and transcriptome analysis during seed development. BMC Genom. 2015;16:208. doi: 10.1186/s12864-015-1429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ning C.L., Jiang Y., Meng J.S., Zhou C.H., Tao J. Herbaceous peony seed oil: A rich source of unsaturated fatty acids and γ-tocopherol. Eur. J. Lipid Sci. Technol. 2015;117:532–542. doi: 10.1002/ejlt.201400212. [DOI] [Google Scholar]
- 19.Biedermann M., Grob K. Comprehensive two-dimensional gas chromatography for characterizing mineral oils in foods and distinguishing them from synthetic hydrocarbons. J. Chromatogr. A. 2015;1375:146–153. doi: 10.1016/j.chroma.2014.11.064. [DOI] [PubMed] [Google Scholar]
- 20.Mravec J., Skupa P., Bailly A., Hoyerova K., Krecek P., Bielach A., Petrasek J., Zhang J., Gaykova V., Stierhof Y.D. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- 21.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Yurchenko O.P., Park S., Ilut D.C., Inmon J.J., Millhollon J.C., Liechty Z., Page J.T., Jenks M.A., Chapman K.D., Udall J.A., et al. Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium. BMC Plant Biol. 2014;14:312. doi: 10.1186/s12870-014-0312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda O., Sakamoto H., Hashimoto T., Iba K. A temperature-sensitive mechanism that regulates post-translational stability of a plastidial omega-3 fatty acid desaturase (FAD8) in Arabidopsis leaf tissues. J. Biol. Chem. 2005;280:3597–3604. doi: 10.1074/jbc.M407226200. [DOI] [PubMed] [Google Scholar]
- 24.Román Á., Andreu V., Hernández M.L., Lagunas B., Picorel R., Martínez-Rivas J.M., Alfonso M. Contribution of the different omega-3 fatty acid desaturase genes to the cold response in soybean. J. Exp. Bot. 2012;63:4973–4982. doi: 10.1093/jxb/ers174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L., Ye J., Guo W.D., Wang C.C., Hu H.T. Differences in cold tolerance and expression of two fatty acid desaturase genes in the leaves between fingered citron and its dwarf mutant. Trees. 2012;26:1193–1201. doi: 10.1007/s00468-012-0695-6. [DOI] [Google Scholar]
- 26.Iba K., Gibson S., Nishiuchi T., Fuse T., Nishimura M., Arondel V., Hugly S., Somerville C. A gene encoding a chloroplast omega-3 fatty acid desaturase complements alterations in fatty acid desaturation and chloroplast copy number of the fad7 mutant of Arabidopsis thaliana. J. Biol. Chem. 1993;268:24099–24105. [PubMed] [Google Scholar]
- 27.Nguyen H.M., Cuiné S., Beyly-Adriano A., Légeret B., Billon E., Auroy P., Beisson F., Peltier G., Li-Beisson Y. The green microalga Chlamydomonas reinhardtii has a single ω-3 fatty acid desaturase that localizes to the chloroplast and impacts both plastidic and extraplastidic membrane lipids. Plant Physiol. 2013;163:914–928. doi: 10.1104/pp.113.223941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H.U., Lee K.R., Shim D., Lee J.H., Chen G.Q., Hwang S. Transcriptome analysis and identification of genes associated with ω-3 fatty acid biosynthesis in Perilla frutescens(L.) var. frutescens. BMC Genom. 2016;17:474. doi: 10.1186/s12864-016-2805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue Y., Chen B., Win A.N., Fu C., Lian J., Liu X., Wang R., Zhang X., Chai Y. Omega-3 fatty acid desaturase gene family from two ω-3 sources, Salvia hispanica and Perilla frutescens: Cloning, characterization and expression. PLoS ONE. 2018;13:e0191432. doi: 10.1371/journal.pone.0191432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyer J.M., Chapital D.C., Cary J.W., Pepperman A.B. Chilling-sensitive, post-transcriptional regulation of a plant fatty acid desaturase expressed in yeast. Biochem. Biophs. Res. Commun. 2001;282:1019–1025. doi: 10.1006/bbrc.2001.4667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.